Abstract
ProfCom is a web-based tool for the functional interpretation of a gene list that was identified to be related by experiments. A trait which makes ProfCom a unique tool is an ability to profile enrichments of not only available Gene Ontology (GO) terms but also of ‘complex functions’. A ‘Complex function’ is constructed as Boolean combination of available GO terms. The complex functions inferred by ProfCom are more specific in comparison to single terms and describe more accurately the functional role of genes. ProfCom provides a user friendly dialog-driven web page submission available for several model organisms and supports most available gene identifiers. In addition, the web service interface allows the submission of any kind of annotation data. ProfCom is freely available at http://webclu.bio.wzw.tum.de/profcom/.
INTRODUCTION
Relating experimental data to biological knowledge is a necessity to cope with the data avalanches emerging from recent developments in high-throughput technologies. Automatic functional profiling has become the de facto approach for the secondary analysis of high-throughput data. A number of tools employing available gene functional annotations as well as pathway databases have been developed (1–18). The advantages and limitations of most of these tools are reviewed in ref. (19).
An important aspect of standard functional profiling methodology is inability to overcome the limits of employed annotation vocabularies. Do current annotation vocabularies cover all possible biological functions? Can they cover them in the future? The space of possible biological functions is almost infinite. However, to control it one does not need an infinite number of functional terms. Consider a very direct analogy. Human language contains a limited number of words but through grammar rules these words can be transformed into an almost infinite number of sentences, which allow the expression of almost any idea. In our previous paper (20), we proposed to construct new functional terms (referred to as ‘complex functions’). A ‘complex function’ is constructed as a combination of available terms. The three Boolean operations (‘AND’, ‘OR’, ‘NOT’) play the role of grammar rules and resulting space of ‘complex functions’ covers an almost infinite number of possible biological functions.
The present article describes ProfCom, a web tool for functional profiling based on the concept described previously (20). ProfCom supports automatic analyses for several model organisms as well as provides a web service interface, which allows the submission of any kind of annotation data. For each organism, ProfCom provides analysis of different annotations, including Gene Ontology (GO) (21), FunCat (22) and InterPro Motifs (23). ProfCom currently offers automatic analyses for Homo sapiens, Mus musculus, Rattus norvegicus, Caenorhabditis elegans, Drosophila melanogaster and Saccharomyces cerevisiae. In addition, any organism and annotation can be analyzed by ProfCom using Web service interface.
MATERIALS AND METHODS
Statistical analysis and ProfCom profiling engine
A standard tool for automatic functional profiling accepts a query list of genes (referred to as set A, usually the set of genes experimentally identified to be related to the studied biological phenomena) and a reference set (referred to as set B, usually the set of all genes from the analyzed organism). Then, for each attribute f from the set F (f is usually a functional term from the employed annotation vocabulary F, i.e. GO, FunCat, etc.) the number af genes in set A and bf genes in set B that have been annotated with f is counted. In the next step, the null hypothesis H0 (genes that belong to the set A are independent of having attribute f) is tested. Hypergeometric, binomial or χ2-tests are usually employed to find over/under represented attributes (19).
Unlike most currently available web tools for functional profiling, ProfCom implements different profiling paradigms. Along with standard profiling of functional terms f (referred to as ‘base’ categories) from annotation vocabularies it also searches for the enrichment related to ‘complex functions’, which are defined as any Boolean combination of ‘base’ categories (for example, a new ‘complex function’ w may define the set of genes that belongs simultaneously to the ‘base’ categories f1 and f2). We consider intersection, union and difference operations. For example, intersection of two categories f1 and f2 is formally defined as ‘complex function’ w = f1 ∩ f2. In other words, w corresponds to the set of genes that belong to both categories f1 and f2. The union of two categories f1 and f2 is formally defined as w = f1 ∪ f2. In this case, w corresponds to the set of genes that belong either to category f1 or f2. The difference between two categories f1 and f2 is formally defined as w = f1/f2; ‘complex function’ w corresponds to the set of genes from category f1 excluding those that simultaneously belong to category f2.
Each ‘complex function’ is characterized by the number of base categories required to construct it. We will refer to this characteristic as degree. For example, the base categories can be defined as ‘complex functions’ of the first degree, the category w = f1 ∩ f2 is a ‘complex function’ of the second degree (intersection).
Consideration of all possible ‘complex functions’ leads to combinatorial complexity. To analyze enrichments for all possible combinations of degree higher than 2 is computationally infeasible. For this reason, a search algorithm should be used. ProfCom employs the algorithm based on greedy heuristics (20). Greedy heuristics does not guarantee to find the optimal solution in every case but significantly reduce the computational complexity. To adjust P-values for multiple testing ProfCom uses the Monte–Carlo simulation approach. The estimated P-value corresponds exactly to the definition of an experiment-wise Westfall and Young P-value (3,20,24). More details on the searching algorithm and P-value adjustment can be found in Supplementary Materials.
Automatically supported annotations and gene Ids
As input ProfCom accepts several types of gene or protein identifiers. For example, for the human genome ProfCom supports identifiers from ‘Entrez Gene’ (25), ‘UniProt/Swiss-Prot’, ‘Gene Symbol’ (25,26), ‘UniGene’ (25), ‘Ensembl’ (27), ‘RefSeq Protein ID’, ‘RefSeq Transcript ID’ (28) and ‘Affymetrix probe codes’ (29). Additionally, a mixture of several identifier types is possible.
In the first step, user-supplied gene Ids are mapped to ‘Entrez Gene’ identifiers. For this purpose, files from NCBI and Affymetrix websites are used. Detailed information on data sources used by ProfCom is in Table 1.
Table 1.
Types of gene identifiers recognized by ProfCom and data sources used for Id mapping
| Type of Ids | File used |
|---|---|
| ‘Gene Symbol’, ‘Ensembl’, ‘LocusTag’ | ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/gene_info.gz |
| ‘RefSeq Protein ID’, ‘RefSeq Transcript ID’ | ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/gene2refseq.gz |
| ‘UniProt/Swiss-Prot’ | ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/gene_refseq_uniprotkb_collab.gz |
| ‘UniGene’ | ftp://ftp.ncbi.nlm.nih.go/gene/DAT/gene2unigene |
| ‘Affymetrix probe codes’ | http://www.affymetrix.com/Annotation files |
The user gets full information on mapping of the supplied gene ids. It includes four tables along with the ProfCom results online. Table 1 reports full mapping details of recognized gene Ids. It includes the informational source used as well as a possible multiple mapping of the user supplied Ids to ‘Entrez Gene’ Ids. Table 2 reports unrecognized gene Ids. Table 3 reports the final mapping (one-to-one mapping), which is used in subsequent analyses. ProfCom implements simple heuristics to resolve multiple mapping issues. If it is possible to map a particular gene Id to several ‘Entrez Gene’ Ids, the Id which has the most abundant annotation is selected. However, if the user finds this mapping to be incorrect (Table 3) he/she can simply resubmit the data by substituting those ambiguous gene Ids with ‘Entrez Gene’ Ids considered to be correct. On the other hand, if several supplied gene Ids are mapped to the same ‘Entrez Gene’ Id then they are considered as belonging to one gene and the Ids are reported concatenated together by a semicolon (‘;’). Table 4 reports all such cases.
Table 2.
Data file used by ProfCom to automatically retrieve annotations
| Annotation | File used |
|---|---|
| Gene Ontology | ftp://ftp.ncbi.nlm.nih.go/gene/DAT/gene2unigene |
| InterPro Motifs | ftp://ftp.ebi.ac.uk/pub/databases/interpro/protein2ipr.dat |
| FunCat | http://mips.gsf.de/ |
We would like to point out that protein and gene identifiers can be highly ambiguous (30) with multiple synonymous variants. For this reason, the quality of the retrieved annotation can be different for different types of identifiers. Several powerful recourses to map different type of gene Ids exist (http://beta.uniprot.org/). To escape multiple mapping issues, we recommend submitting ‘Entrez Gene’ identifies to ProfCom.
ProfCom automatically supports several annotations. Currently, they include GO (21), FunCat (22) and InterPro Motifs (23). Detailed information on data sources used to retrieve each annotation is presented in the Table 2. The ProfCom web interface allows the user to use all annotations simultaneously or combine them.
In addition to the interactive web-submissions, custom annotation data can be analyzed using the ProfCom Web service. This allows the use of ProfCom for almost any problem domain, e.g. different annotation types or organisms. Furthermore, web services enable one to run ProfCom analyses in pipelines or automated workflows from most systems. This ensures a fast and convenient usage for a broad range of use cases: starting from a quick hypothesis evaluation to detailed high-quality annotations.
Implementation
ProfCom runs on a standard Apache/Tomcat web server. The actual profiling algorithm is implemented in Java and C for platform independence and high performance. The computation is distributed on Linux workstations utilizing a Sun Grid engine and thus ensures scalability. A ProfCom analysis starts by user-friendly dialog-driven web form. In the first step, the model organism is chosen and the list of gene or protein names of interest is uploaded. Optionally, the reference set of genes can be uploaded. By default, the set of all annotated genes (‘Entrez Gene’ Ids) from the chosen organism is used as the reference set. Depending on the chosen organism the ProfCom web page automatically shows all available annotations.
Illustration of ProfCom model inference process
Here, we present one example of analyses of real data by ProfCom to illustrate it novelties and utilities in comparison to existing related tools. More examples can be found in Supplementary Materials, where we bring together several independent studies that performed gene expression analyses to identify over/under expressed genes in different cancer types. We collect a set of differentially expressed genes originally identified in each study (we refer to each of these sets as set A and the set of all human genes is referred to as set B).
In ref. (31), microarray experiments were done to compare gene expression in 50 ovarian cancer specimens, including all four histotypes to gene expression in five pools of normal ovarian surface epithelial cells. Data were analyzed to determine whether changes in gene expression correlated with different histotypes, grade or stage.
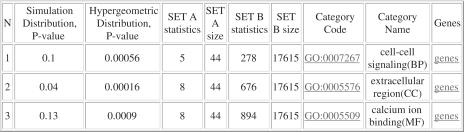
Several set of genes that show the greatest ability to differentiate between considered cancer subtypes were originally identified. For example, 47 selected genes were 2-fold differentially expressed in mucinous ovarian cancers compared to other histotypes and with normal ovarian surface epithelial cells. Standard functional profiling reveals several GO term significantly overrepresented. It is widely known that the processes of Ca++ homeostasis are often disordered in many cancer types (32). Therefore, the presence of GO term ‘calcium-ion binding’ among top enriched GO terms is of particular interest. Eight genes (MRC1, EFHD2, PLS1, ANXA10, LDLR, MMP1, S100P, THBS2) from the set A are related by this term (Figure 1). On the other hand, there are 894 genes in the whole human genome classified as ‘calcium-ion binding’. Using conventional GO terms vocabularies, standard profiling procedure is not able to supply evidences that would discriminate these eight genes (from all human 894 ‘calcium-ion binding’) and, thus, to clarify molecular mechanism involved.
Figure 1.
ProfCom output table ‘Top enriched categories of degree 1’ for the considered example.
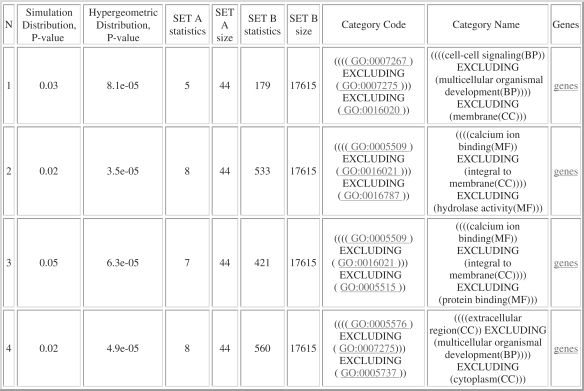
The complex function ‘calcium-ion binding EXCLUDING integral to membrane EXCLUDING hydrolase activity’ inferred by ProfCom (Figure 2) relates all ‘calcium-ion binding’ genes from the set A and is more specific in comparison to a single GO term, i.e. only 533 genes (compared to 894) in the human genome are classified by this complex function. It is not only better from statistical viewpoint (equal selectivity with ∼1-fold increase in specificity), but also supplies valuable biological information which can be helpful for making biological conclusions about molecular mechanisms involved in the considered cancer type.
Figure 2.
ProfCom output table ‘Top enriched categories of degree 3’ for the considered example.
CONCLUSION
Automatic functional profiling becomes the de facto approach for the secondary analysis of high-throughput data. A number of tools employing available gene functional annotations have been developed. However, most of these tools are limited by available annotation vocabularies and may fail to provide full interpretation of biological relationships in a set of genes involved in complex biological phenomena. Here, we present ProfCom, a web-based tool that implements the new profiling paradigm for the interpretation of functional relations between genes. ProfCom profiling engine employs three logical operations (‘AND’, ‘OR’, ‘NOT’) to provide complex functions that classify more specifically the biological role of a gene group.
As been demonstrated, in many cases, complex functions provide better understanding of molecular mechanisms involved for the phenomena under study. On the other hand, in some cases, relative GO terms can form many redundant complex functions and may complicate the manual analyses of the ProfCom results. This may be considered as a potential disadvantage. One potential way to resolve redundancy problem is the inclusion of methodologies that group related sets of annotations before the analyses (18,33,34), in the future.
ProfCom provides technical support to the user that corresponds to the best currently available standards in the field. It has a dialog-driven web page for submission that covers several mostly exploited model organisms. In addition, the web service interface allows one submitting any kind of annotation data and is not limited to a particular organism or problem domain. This property significantly simplifies the procedure of data analyses and increases the spectrum of gene sets that can be analyzed. These features make ProfCom an attractive practical tool for biologists interpreting new experimental data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Ulrich Gueldener, Philip Wong for helpful discussions and Michael Strasser for initial technical help at the project beginning. T.S. was supported by the DFG Program ‘Bioinformatics Initiative Munich’. Funding to pay the Open Access publication charges for this article was provided by Helmholtz Zentrum Munich, German Research Center for Environmental Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Antonov AV, Mewes HW. BIOREL: the benchmark resource to estimate the relevance of the gene networks. FEBS Lett. 2006;580:844–848. doi: 10.1016/j.febslet.2005.12.101. [DOI] [PubMed] [Google Scholar]
- 2.Antonov AV, Tetko IV, Mewes HW. A systematic approach to infer biological relevance and biases of gene network structures. Nucleic Acids Res. 2006;34:e6. doi: 10.1093/nar/gnj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berriz GF, King OD, Bryant B, Sander C, Roth FP. Characterizing gene sets with FuncAssociate. Bioinformatics. 2003;19:2502–2504. doi: 10.1093/bioinformatics/btg363. [DOI] [PubMed] [Google Scholar]
- 4.Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007;8:R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khatri P, Draghici S, Ostermeier GC, Krawetz SA. Profiling gene expression using onto-express. Genomics. 2002;79:266–270. doi: 10.1006/geno.2002.6698. [DOI] [PubMed] [Google Scholar]
- 7.Khatri P, Bhavsar P, Bawa G, Draghici S. Onto-Tools: an ensemble of web-accessible, ontology-based tools for the functional design and interpretation of high-throughput gene expression experiments. Nucleic Acids Res. 2004;32:W449–W456. doi: 10.1093/nar/gkh409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khatri P, Voichita C, Kattan K, Ansari N, Khatri A, Georgescu C, Tarca AL, Draghici S. Onto-Tools: new additions and improvements in 2006. Nucleic Acids Res. 2007;35:W206–W211. doi: 10.1093/nar/gkm327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin D, Brun C, Remy E, Mouren P, Thieffry D, Jacq B. GOToolBox: functional analysis of gene datasets based on Gene Ontology. Genome Biol. 2004;5:R101. doi: 10.1186/gb-2004-5-12-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masseroli M, Martucci D, Pinciroli F. GFINDer: genome function integrated discoverer through dynamic annotation, statistical analysis, and mining. Nucleic Acids Res. 2004;32:W293–W300. doi: 10.1093/nar/gkh432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, Narasimhan S, Kane DW, Reinhold WC, Lababidi S, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Shahrour F, az-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 14.Al-Shahrour F, Minguez P, Vaquerizas JM, Conde L, Dopazo J. BABELOMICS: a suite of web tools for functional annotation and analysis of groups of genes in high-throughput experiments. Nucleic Acids Res. 2005;33:W460–W464. doi: 10.1093/nar/gki456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Shahrour F, Minguez P, Tarraga J, Montaner D, Alloza E, Vaquerizas JM, Conde L, Blaschke C, Vera J, Dopazo J. BABELOMICS: a systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res. 2006;34:W472–W476. doi: 10.1093/nar/gkl172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Shahrour F, Minguez P, Tarraga J, Medina I, Alloza E, Montaner D, Dopazo J. FatiGO +: a functional profiling tool for genomic data. Integration of functional annotation, regulatory motifs and interaction data with microarray experiments. Nucleic Acids Res. 2007;35:W91–W96. doi: 10.1093/nar/gkm260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goffard N, Weiller G. PathExpress: a web-based tool to identify relevant pathways in gene expression data. Nucleic Acids Res. 2007;35:W176–W181. doi: 10.1093/nar/gkm261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35:W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khatri P, Draghici S. Ontological analysis of gene expression data: current tools, limitations, and open problems. Bioinformatics. 2005;21:3587–3595. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antonov AV, Mewes HW. Complex functionality of gene groups identified from high-throughput data. J. Mol. Biol. 2006;363:289–296. doi: 10.1016/j.jmb.2006.07.062. [DOI] [PubMed] [Google Scholar]
- 21.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mewes HW, Amid C, Arnold R, Frishman D, Guldener U, Mannhaupt G, Munsterkotter M, Pagel P, Strack N, Stumpflen V, et al. MIPS: analysis and annotation of proteins from whole genomes. Nucleic Acids Res. 2004;32:D41–D44. doi: 10.1093/nar/gkh092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD, et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westfall PN, Young SS. New York: John Wiley & Sons; 1993. Resampling-based Multiple Testing: Examples and Methods for P-value Adjustment. [Google Scholar]
- 25.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Birney E, Andrews D, Caccamo M, Chen Y, Clarke L, Coates G, Cox T, Cunningham F, Curwen V, Cutts T, et al. Ensembl 2006. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu G, Loraine AE, Shigeta R, Cline M, Cheng J, Valmeekam V, Sun S, Kulp D, Siani-Rose MA. NetAffx: Affymetrix probesets and annotations. Nucleic Acids Res. 2003;31:82–86. doi: 10.1093/nar/gkg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Draghici S, Sellamuthu S, Khatri P. Babel's tower revisited: a universal resource for cross-referencing across annotation databases. Bioinformatics. 2006;22:2934–2939. doi: 10.1093/bioinformatics/btl372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marquez RT, Baggerly KA, Patterson AP, Liu J, Broaddus R, Frumovitz M, Atkinson EN, Smith DI, Hartmann L, Fishman D, et al. Patterns of gene expression in different histotypes of epithelial ovarian cancer correlate with those in normal fallopian tube, endometrium, and colon. Clin. Cancer Res. 2005;11:6116–6126. doi: 10.1158/1078-0432.CCR-04-2509. [DOI] [PubMed] [Google Scholar]
- 32.Revankar CM, Advani SH, Naik NR. Altered Ca2+ homeostasis in polymorphonuclear leukocytes from chronic myeloid leukaemia patients. Mol. Cancer. 2006;5:65. doi: 10.1186/1476-4598-5-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 34.Grossmann S, Bauer S, Robinson PN, Vingron M. Improved detection of overrepresentation of Gene-Ontology annotations with parent child analysis. Bioinformatics. 2007;23:3024–3031. doi: 10.1093/bioinformatics/btm440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.