Abstract
One of the challenges of the post-genomic era is to provide accurate function annotations for large volumes of data resulting from genome sequencing projects. Most function prediction servers utilize methods that transfer existing database annotations between orthologous sequences. In contrast, there are few methods that are independent of homology and can annotate distant and orphan protein sequences. The FFPred server adopts a machine-learning approach to perform function prediction in protein feature space using feature characteristics predicted from amino acid sequence. The features are scanned against a library of support vector machines representing over 300 Gene Ontology (GO) classes and probabilistic confidence scores returned for each annotation term. The GO term library has been modelled on human protein annotations; however, benchmark performance testing showed robust performance across higher eukaryotes. FFPred offers important advantages over traditional function prediction servers in its ability to annotate distant homologues and orphan protein sequences, and achieves greater coverage and classification accuracy than other feature-based prediction servers. A user may upload an amino acid and receive annotation predictions via email. Feature information is provided as easy to interpret graphics displayed on the sequence of interest, allowing for back-interpretation of the associations between features and function classes.
INTRODUCTION
Computational approaches to protein annotation prediction often infer protein function by transferring annotations between proteins with similar sequence, structure, amino acid motifs or phylogenetic profiles. Most automated function prediction servers employ nearest neighbour approaches that rely upon identifying well-annotated sequence and structural homologues. In practice, these methods are only applicable in cases where sequence relationships can be reliably established and homologues are functionally well characterized. Past estimates based on 2 million known sequences suggested as few as 33% of unannotated sequences were closely related to well-characterized homologues and could be targeted by these methods (1). The remaining set comprised sequences that were distantly homologous to well-annotated proteins or were orphan proteins. More recent studies have shown that similar sequences cannot always be used to infer similar functions, for example, one study reported a requirement of 40–70% sequence identity between enzymes to transfer function with 90% accuracy (2). A second study highlighted the problems of ‘annotation lag’ or indeed erroneous annotations in sequence databases. Often, proteins that had been well characterized experimentally still exist as ‘hypothetical proteins’ within the biological sequence databases over 2 years after the original literature has been published characterizing the sequence (3). These findings highlight the importance for accurate automated methods that can be applied to all sequences and are independent of homology information.
One class of method that addresses the annotation of orphan and unannotated proteins are feature-based. These methods utilize information derived from characteristics of the protein sequence; secondary structure or hydrophobicity for example, in order to determine function. Machine-learning feature-based approaches have been successfully used to recognize patterns of features that are indicative of different functional classes such as enzyme EC numbers (4) and for a handful of Gene Ontology (GO) terms (5). These methods do not rely on annotation transfer from nearest neighbour sequences and are resistant to missing or error-prone annotations through the use of sensitive machine learning techniques that are capable of distinguishing genuine functional signals from noise. Consequently, these approaches are capable of providing truly novel functional insights by generating ab initio function predictions.
Here, we describe the FFPred server for feature-based function prediction using the Gene Ontology Annotations (6) as our definition of function. This server differs from other conventional function prediction servers in that it has the capacity to annotate orphan and distantly homologous proteins with broad function terms. The server consists of individual classifiers for 111 molecular function and 86 biological process categories capable of achieving >50% sensitivity at false positive rates of <10%. The categories represent over 300 GO annotation terms considering inheritance within the term definitions. In a comparison study with another feature-based server (7), FFPred offered the broadest selection of GO annotation terms for prediction and achieved a greater level of accuracy for common classifiers through the use of additional feature inputs and more accurate prediction of existing features using PSI-BLAST profiles.
METHODS
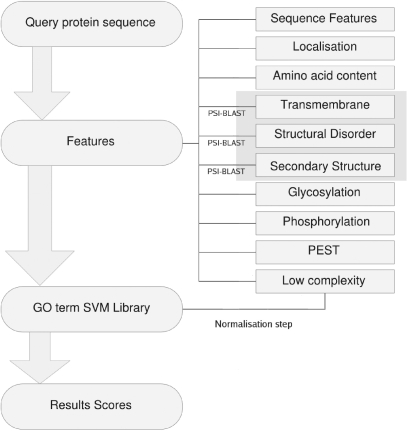
The server processing model (Figure 1) shows the computational steps involved from inputting a query amino acid sequence to generating a set of GO term predictions. The first step involves the generation of a set of feature descriptors for the query sequence. The features are calculated from a suite of programs predicting cellular localization, post-translational modification patterns, secondary structure and transmembrane regions [see ref. (7) for full feature listing]. Most of the prediction algorithms require a single amino acid sequence as input; however, for secondary structure, disorder and transmembrane features, more accurate predictions can be obtained by the use of PSI-BLAST profiles as input to the algorithms. For this purpose, three separate PSI-BLAST profiles are generated according to the recommended default parameter settings for each prediction program using the current version of uniref90 (8) as the search database.
Figure 1.
Server process flow diagram.
Each program output is parsed and converted into feature descriptors describing attributes of the sequence such as number of predicted disordered residues at the N-terminus, or number of glycosylated residues predicted in the protein. At this stage, the feature matrix is normalized so that values lie between 0 and 1 and reformatted for screening against the library of support vector machines (SVMs). Each classifier outputs a binary decision value as to whether the protein should receive the annotation term or not, with an associated posterior probability. The decision values and probabilities are collated for each GO term and summarized in the final annotation results as jury decisions and confidence scores.
Each GO class is represented by five SVM classifiers trained with rbf (radial basis function) kernels to recognize feature patterns associated with the annotation term. The rbf kernel was selected due to its simplicity and superior performance when applied to a variety of biological problems (9,10). The rbf kernel has also been shown to be a general case of linear kernel and sigmoid kernel for certain parameters (11). SVMs for a given category were trained using five homology reduced partitions of the total 14 055 protein dataset. The number of partitions was selected as a trade-off between retaining sufficient positive class examples for model building and the ability to predict an annotation using multiple classifiers. Sequences in the same training and test dataset partition were filtered so that no two sequences were related at more than 1e-6 BLAST E-value. This technique boosted overall performance by providing five independent classifiers utilizing different feature weightings for prediction of the same GO category.
GO class scoring scheme
The posterior probabilities for each classifier are generated using the method of Platt (12). In this case, the SVM model encodes the position of a decision boundary separating the positive GO class members from the negative GO class members according to the feature input data. The distance from the decision boundary in either direction for a protein represented in feature space can be obtained from the classifier output. Probabilities are assigned to each distance value f(x) for a GO classifier y by estimating the parameters A and B of a sigmoid function that is fitted to the distribution of distances obtained for an independent test protein dataset (Equation 1).
| 1 |
A GO class assignment is made by majority rule for a protein, where three out of the five classifiers produce a positive decision result. This number is reported as the Jury value (Figure 2). The final confidence score for a GO class assignment is taken as the average of the three highest probability values. As a guide, the greater the number of classifiers that predict an annotation class for a protein (the Jury value), the greater the confidence in the resulting score. This scoring mechanism eliminates false positive assignments made by one or two of the five classifiers increasing overall precision.
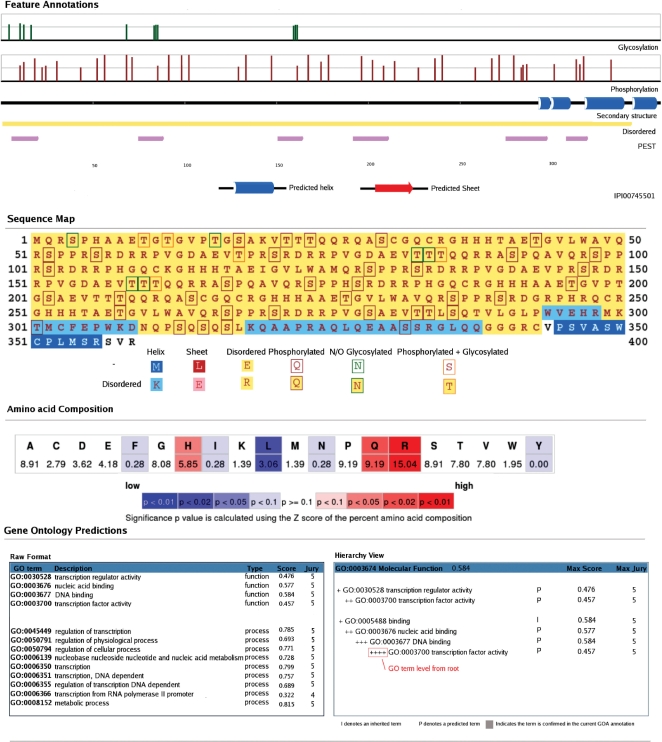
Figure 2.
Sample output from the server for sequence IPI00745501.
Finally, the hierarchical nature of the GO classification scheme is exploited by propagating annotation confidence scores between related terms. A maximum confidence score is reported and displayed in the GO hierarchy view (Figure 2) for each GO term representing the maximum confidence score of the term classifier or any of its child terms.
USAGE
Input query format
The FFPred server accepts single protein sequences as input formatted as plain text or in FASTA format. It is expected that the amino acid sequence of interest represents the entire mature protein product of a gene or at least a genuine transcript. Server results based on sequence fragment inputs may be unreliable as feature information may differ substantially between truncated gene products. Additionally, if the sequence input has been recently processed or is present in the human IPI protein dataset, the user will be immediately directed to a page displaying the feature and GO term predictions for the given query sequence.
Database query
The user may also view pre-processed results for the human proteome using IPI accession identifiers or enter one of the GO terms in the library into the GO query field. In the latter case, the reported results table details novel predictions that are not present within the GOA annotation or Swiss-Prot annotated datasets.
Output format
Server output for sequence submissions are returned to the user by email containing a text summary of GO annotation predictions for the input sequence with a hyperlink to a dynamically generated temporary results page (Figure 2). The results page details predicted features and GO annotations for the query sequence. The feature predictions are shown in tabular format as well as graphically mapped onto the sequence of interest for easy interpretation. This allows for back interpretation of feature patterns responsible for functions. This view is also available in print friendly format.
GO term predictions are represented in hierarchical format or as single table of individual term results. In the hierarchy view, each GO term is annotated according to whether it was predicted by classifiers present in the library, or whether the annotation was inherited through classifiers representing one or more of the child terms. This view enables the user to contextualize the predictions and derive extra confidence in predictions that are made by both parent and child term classifiers.
Computational efficiency
In the case of a typical protein sequence, computation takes 12–15 min from initial sequence submission to receiving server results via email on an Intel Xeon 3.2 GHz processor running CentOS 4.4. The majority portion of this time is spent screening the GO term SVM library (around 11 min per sequence). Users wishing to submit significant number of queries or whole proteomes for annotation should contact the authors for advice.
RESULTS AND DISCUSSION
The SVM models underlying this method have been trained and tested on human annotated proteins. In order to assess the performance of the method on other organisms, we tested the classifiers using Gene Ontology Annotations from the GOA project (13) on eukaryotic model organisms zebrafish (Danio rerio), mouse (Mus musculus), fly (Drosophila melanogaster), worm (Caenorhabditis elegans) and yeast (Saccharomyces cerevisiae). Table 1 lists the performance statistics; sensitivity, specificity, precision and Matthew's correlation coefficient (MCC) obtained for each organism using the classifiers trained on human data for all categories performing better than random. The proteins in each genome that were annotated with one or more GO terms were used as the basis of the benchmark study. A result was considered correct if the server assignment was also represented in the GOA annotation by the GO term in question or one of its child terms. Proteins annotated at less specific GO term levels than the term in question were omitted from the study.
Table 1.
Classification performance for six eukaryotic proteomes
| MCC | Sensitivity | Specificity | Precision | No. of Proteins | No. of Categories | |
|---|---|---|---|---|---|---|
| Human | 0.66 | 0.67 | 0.99 | 0.68 | 32 528 | 197 |
| Mouse | 0.57 | 0.48 | 0.98 | 0.52 | 26 557 | 196 |
| Zebrafish | 0.65 | 0.58 | 0.97 | 0.64 | 12 684 | 186 |
| Worm | 0.47 | 0.47 | 0.97 | 0.56 | 11 770 | 165 |
| Fly | 0.44 | 0.40 | 0.98 | 0.57 | 13 107 | 175 |
| Yeast | 0.42 | 0.34 | 0.97 | 0.61 | 5527 | 99 |
Each performance statistic represents the mean average value for all GO term classifiers. MCC represents Matthew's correlation coefficient, a measure of overall classifier accuracy. A value of 0 indicates random performance, whilst a value of 1 implies perfect classification. Sensitivity represents the proportion of positive examples recovered by the classifier, i.e. TP/(TP + FN). Specificity represents the proportion of negatives examples recovered by the classifier i.e. TN/(FP + TN). Precision represents the proportion of positive assignments made by the classifier that were correct, i.e. TP/(TP + FP). TP, true positives; TN; true negatives; FP, false positives; FN, false negatives.
As evolutionary distance between the different species and human increased, the overall average classifier accuracy decreased (MCC values in Table1). Inspection of the sensitivity and specificity values showed the performance decrease could be attributed to a loss in sensitivity across more distantly related species worm, fly and yeast. The sensitivities obtained for mouse and zebrafish were comparable with human. The average specificities observed for all classifiers for each proteome were high for all organisms. This property is a requirement for predictors that will be applied to whole proteomes to avoid large numbers of false positives, where the expected number of GO term annotations is small compared with the number of proteins not annotated by the GO term.
The number of classifiers obtaining over 90% specificity at sensitivities of >30% were also reported (Table1). The decrease in these numbers with evolutionary distance from human can be explained in part as a consequence of differences in quality and completeness between the various proteome annotation efforts and in part as a function of decreasing feature conservation between proteins from more distant eukaryotic proteomes. Amongst the 99 categories that were useful in predicting the functions of yeast proteins, the majority were more general annotation terms that had higher performance accuracies on human proteins and were focused around enzymatic and transmembrane protein functions. The majority of terms unsuitable for use with yeast were biological process categories. This observation suggests that the features corresponding with many of these categories in human are not conserved within lower eukaryotes and may correspond with other studies reporting a lack of conservation of protein–protein interactions between species (14).
Overall, the benchmark results show robust classification accuracies across the vertebrate and mammalian proteomes for most of the annotation categories. We recommend the effective use of this server to annotate vertebrate and mammalian proteomes; however, our results indicate that when run on proteins from lower eukaryotic organisms, the server is more likely to leave a protein unannotated rather than produce an erroneous annotation. The server is not recommended for use with proteins from plants or prokaryotic organisms. Key differences in subcellular localization signalling pathways and post-translational modification pathways mean that patterns of features corresponding with function are not sufficiently conserved with those obtained for human for effective function prediction.
The two primary uses of the server are in the annotation of orphan and unannotated proteins or for partially annotated proteins. Example output (Figure 2) represents the predicted functions for the human IPI00745501 protein sequence. This sequence does not return any annotated hits by sequence homology searches and is therefore unlikely to be annotated by servers utilizing annotation transfer methods. FFPred annotates this sequence with several GO terms with a maximum annotation score of 0.584 as a DNA-binding protein and more specifically a transcription factor. From the feature-based graphical output, we also learn that this sequence contains many predicted phosphorylation and O-glycosylation sites and is compositionally biased with significantly over-represented in arginine, glutamine and histidine residues (red highlights in Figure 2). The sequence is under-represented in leucine, isoleucine and phenylalanine residues (blue highlight in Figure 2) and the predicted subcellular localization is nuclear. These pieces of information can be used to rationalize the prediction, since the presence of many phosphorylation sites within the protein is consistent with a role in some signalling pathway and the large contiguous stretches of disorder enriched in positively charged residues are characteristics of some DNA-binding proteins (15). These types of predictions made on unannotated sequences await further characterization and experimental validation in the laboratory. For predictions made on well-characterized sequences, supporting evidence can often be found in the literature or by comparing FFPred results with other independent prediction methods.
CONCLUSIONS
Genome sequencing projects have furthered our understanding of disease processes and the biological mechanisms underlying them. Sequences with high homology to closely related organisms can be readily annotated by numerous similarity search techniques. As a result, existing function annotations that can be transferred in this way are quickly propagated throughout the biological sequence databases. However, it is clear that many functions are not simply determined by sequence homologies and in many cases we cannot confidently identify relationships with well-characterized proteins. The FFPred server that integrates information from many different resources provides a powerful and necessary alternative to homology inference-based methods and can deliver vital functional clues where other methods fail.
FUTURE WORK
At the time of writing, due to computational costs, the current version of the server did not permit batch sequence processing. We intend to Grid enable the server in the future and anticipate that this feature will be incorporated in future server releases so that users can submit multiple query sequences to the FFPred server.
ACKNOWLEDGEMENTS
We would like to acknowledge M. Sadowski, M. Pentony, Y. Edwards, R. Myers and S. Shah for helpful advice in server design and testing. This work was funded by a BBSRC Doctoral Training Grant with industry sponsorship from BioFocus DPI, and the European Commission within its FP6 Programme, under the thematic are ‘Life sciences genomic and biotechnology for health’ contract LHSG-CT-2003-503265 (BioSapiens Network of Excellence). Funding to pay the Open Access publication charges for this article is provided by the BioSapiens Network of Excellence.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ofran Y, Punta M, Schneider R, Rost B. Beyond annotation transfer by homology: novel protein function prediction methods to assist drug discovery. Drug Discov. Today. 2005;10:1475–1482. doi: 10.1016/S1359-6446(05)03621-4. [DOI] [PubMed] [Google Scholar]
- 2.Rost B. Enzyme function less conserved than anticipated. J. Mol. Biol. 2002;318:595–608. doi: 10.1016/S0022-2836(02)00016-5. [DOI] [PubMed] [Google Scholar]
- 3.Tian W, Skolnick J. How well is enzyme function conserved as a function of pairwise sequence identity? 2003;333:863–882. doi: 10.1016/j.jmb.2003.08.057. [DOI] [PubMed] [Google Scholar]
- 4.Dobson PD, Doig AJ. Predicting enzyme class from protein structure without alignments. 2004;345:187–199. doi: 10.1016/j.jmb.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 5.Jensen LJ, Stærfeldt H-H, Brunak S. Prediction of human protein function according to Gene Ontology categories. Bioinformatics. 2003;19:635–642. doi: 10.1093/bioinformatics/btg036. [DOI] [PubMed] [Google Scholar]
- 6.Asburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lobley A, Swindells MB, Orengo CA, Jones DT. Inferring patterns of native disorder in proteins. PLoS Comp. Biol. 2007;3:e162. doi: 10.1371/journal.pcbi.0030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pirooznia M, Deng Y. SVM classifier – a comprehensive java interface for support vector machine classification of microarray data. BMC Bioinform. 2006;12 doi: 10.1186/1471-2105-7-S4-S25. 7, 4:S25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez M, Caballero J, Fernandez L, Abreu JL, Acosta G. Classification of conformational stability of protein mutants from 3D psuedo-folding graph representation of proteins sequences using support vector machines. Proteins. 2008;70:167–175. doi: 10.1002/prot.21524. [DOI] [PubMed] [Google Scholar]
- 11.Sathiya KS, Lin C-J. Asymptotic behaviours of support vector machines with Gaussian kernel. Neural Comp. 2003;15:1667–1689. doi: 10.1162/089976603321891855. [DOI] [PubMed] [Google Scholar]
- 12.Lin H-T, Lin C-J, Weng RC. Technical report. Department of Computer Science and Information Engineering, National Taiwan University; 2003. A note on Platt's probabilistic outputs for support vector machines. [Google Scholar]
- 13.Camon E, Magrane M, Barrell D, Lee V, Dimmer E, Maslen J. The Gene Ontology Annotation (GOA) Database: sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 2004;32:D262–D266. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mika S, Rost B. Protein-protein interactions more conserved within than across species. PLoS Comp. Biol. 2007;2:e79. doi: 10.1371/journal.pcbi.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Churchill ME, Travers AA. Protein motifs that recognize structural features of DNA. Trends Biochem. Sci. 1991;16:92–97. doi: 10.1016/0968-0004(91)90040-3. [DOI] [PubMed] [Google Scholar]