Abstract
NetMHC-3.0 is trained on a large number of quantitative peptide data using both affinity data from the Immune Epitope Database and Analysis Resource (IEDB) and elution data from SYFPEITHI. The method generates high-accuracy predictions of major histocompatibility complex (MHC): peptide binding. The predictions are based on artificial neural networks trained on data from 55 MHC alleles (43 Human and 12 non-human), and position-specific scoring matrices (PSSMs) for additional 67 HLA alleles. As only the MHC class I prediction server is available, predictions are possible for peptides of length 8–11 for all 122 alleles. artificial neural network predictions are given as actual IC50 values whereas PSSM predictions are given as a log-odds likelihood scores. The output is optionally available as download for easy post-processing. The training method underlying the server is the best available, and has been used to predict possible MHC-binding peptides in a series of pathogen viral proteomes including SARS, Influenza and HIV, resulting in an average of 75–80% confirmed MHC binders. Here, the performance is further validated and benchmarked using a large set of newly published affinity data, non-redundant to the training set. The server is free of use and available at: http://www.cbs.dtu.dk/services/NetMHC.
INTRODUCTION
Intracellular infections with pathogens such as viruses and certain bacteria are defeated by cytotoxic T lymphocytes (CTL). The CTL T-cell receptor (TCR) recognizes foreign peptides in complex with major histocompatibility complex (MHC) class I molecules on the surface of the infected cells. MHC class I molecules preferably bind and present nine amino acid long peptides, which mainly originates from proteins expressed in the cytosol of the presenting cell. In most vertebrates, MHCs exist in a number of different allelic variants that each binds a specific and very limited set of peptides. For a number of years, prediction methods have developed to identify which peptides will bind a given MHC (1), and such predictions can be highly valuable in a broad range of applications, including rational vaccine design and disease diagnostics. The artificial neural network (ANN) training method behind NetMHC (2,3) has been benchmarked to be the best among available methods (4). Preliminary versions of the algorithm have been used to predict possible MHC-binding peptides in a large set of pathogenic viral proteomes, resulting in an average of >75% confirmed MHC binders (5). Most MHC prediction algorithms (a list of other servers is included in the Supplementary Material) are trained on peptides of the same length as they predict, but since data for peptide lengths different from nine are much more scarce, the broadness of MHC binding predictions for different peptide lengths is accordingly limited. In this server, however, a method is implemented making it possible to predict 8-, 10- and 11-mer peptide binding using 9-mer trained predictors, which extends the MHC coverage for these peptide lengths significantly compared to other available MHC:peptide-binding servers.
METHODS
The server is trained on the largest number of quantitative peptide:MHC affinity measurements ever published using both affinity data from the Immune Epitope Database and Analysis Resource (IEDB) (6), eluted peptide data from the SYFPEITHI database (7) and proprietary affinity data. The predictions based on ANNs are trained essentially as described in (3) on data from 55 MHC alleles (43 Human and 12 non-human), and the predictions based on position specific scoring matrices (PSSMs) are trained as described in (2) for additional 67 HLA alleles. A large number of 9-mer MHC affinity data have become available from the IEDB database, since the training of the ANNs used at NetMHC-3.0, and all peptides not used in the training (6452 9-mer peptide affinity data points, covering 32 HLA alleles) were used for evaluation of the server performance. These data are available at the server. In this dataset, 3104 were measured to be binders (IC50<500 nM), 76% of these were correctly predicted as such. 3030 peptides were predicted to bind to a given HLA, and 78% of these had a measured IC50<500 nM. The average Pearson correlation coefficient (PCC) and area under a ROC curve (AUC) value using a 500 nM classification threshold were 0.71 and 0.86, respectively. For the full per allele results, see the Supplementary Material (Supplementary Table 1 and Supplementary Figure 1). NetMHC-3.0 uses a new approximation algorithm that reliably predicts the affinity of peptides of lengths 8, 10 and 11, for which affinity data for training are rare (8). The method uses predictors trained on peptides of length 9 to successfully extrapolate to other lengths. In short, the method approximates each peptide of any length to a number of 9-mers, by inserting X (for 8-mers) or deleting amino acid(s) (for 10- and 11-mers) and set the final prediction to an average of the 9-mer predictions. We had previously trained ANN predictors directly on 10-mer affinity data and since this training more than 2000 10-mer peptide:MHC affinities had become available from the IEDB database (6). Area under a ROC curve (AUC) values were calculated for each allele using either ANNs trained on 10-mers or the approximation method. For 12 of the 16 alleles, the approximation method performed better than the 10-mer trained ANNs (P < 0.01), see Supplementary Material Figure 2. However, for the four HLA-alleles, this evaluation showed better performance for ANNs trained on 10-mer peptides; these 10-mer trained ANNs are used for predictions by the server. For 8-mers, 2002 affinity data were extracted covering 35 MHC alleles. The overall PCC and AUC were 0.68 and 0.86, respectively. For 8-mer per allele performance, see the Supplementary Material Figure 4. For 8-mers, predictors trained on actual 8-mers seems to be better than the approximation method otherwise used, so for the alleles with available 8-mer affinity data, 8-mer trained ANNs are used for the predictions. In general, it is not possible to estimate how reliable a single prediction is. However, the stronger the affinity is predicted the higher are the chance that the actual affinity is stronger than the generally accepted binding threshold of 500 nM.
SERVER
NetMHC-3.0 predicts the binding affinity of either a list of peptides with a defined length (8–11 residues) or all possible sub-peptides hosted within full-length proteins. The input must be in the FASTA format, or as peptides all of equal length, one peptide pr. line. The server will accept a maximum of 5000 sequences per submission; each sequence not more than 20 000 amino acids with a minimum length corresponding to the selected length of prediction (see subsequently). Input data can be pasted into a text field or uploaded from a local file on the user's computer.
If the input is in peptide, format the corresponding tick-box must be selected. The input must not exceed 5000 sequences and with a maximum of 20 000 amino acids in each sequence. One or more MHCs must be selected, as well as the desired peptide length. Only one prediction length at a time can be used. The output can optionally be sorted according to the predicted affinity by selecting a tick-box. The predictions start by clicking the Submit button. An example input in FASTA format is shown in Figure 1.
Figure 1.
Example input in FASTA format.
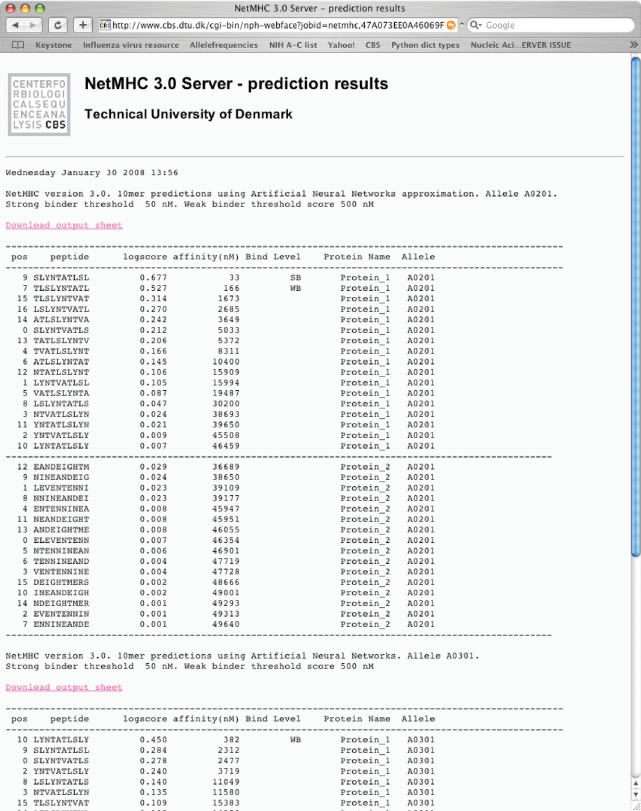
The output is displayed as raw text with a header indicating the server name, the type of prediction (PSSM, ANN or ANN-approximation) the first selected allele and the date (Figure 2) followed by the prediction output in a column format. The columns are named in the first line of the prediction output. The first column [pos] is the position of the first amino acid of the predicted peptide within the possibly longer sequence, numbering starting with 0. Column (peptide) is the primary sequence of the (sub-)peptide. Column (logscore) is the raw prediction output, which for ANNs is 1-log50000 to the affinity in nanomolar units. For PSSM predictions the raw prediction score is a log-odds likelihood score. Additionally a column is included for ANN predictions, [affinity (nM)], which is the predicted affinity presented in nanomolar units. Column (Bind Level) indicates if the peptide is predicted to bind stronger than a certain threshold [for ANN predictions stronger than 50 nM (SB) or stronger than 500 nM (WB); for PSSM high-binding peptides (SB) have a prediction score greater than the 0.1% percentile score value of 1 000 000 random natural peptides, and weak binding (WB) peptides a score value above the 1% percentile score of 1 000 000 random natural peptides predictions]. Predicted affinities weaker than 500 nM or lower than the 1% percentile score have no indications. Column (Protein Name) gives the name of the predicted protein. If peptide input was used, the name will always be ‘Sequence’. Column (Allele) gives the name of the MHC allele chosen. The output contains all the sub-peptides for each protein for a given allele either in the order they appear in the sequence or sorted by predicted affinity within each protein (if chosen). If more than one protein sequence were entered, a dashed line will separate the peptides from each protein. If more than one allele were chosen, the output will show a header similar to the first immediately after the first predictions, all in the same web output page.
Figure 2.
Raw text output using the input in Figure 1 and selecting the alleles HLA-A0201 and HLA-A0301. 10-mer peptide predictions were chosen. Affinity sorting was chosen.
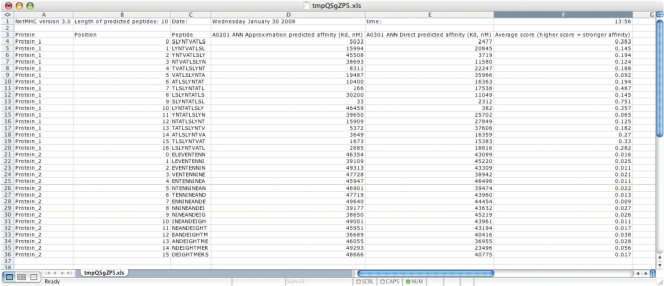
In each header, there is a link to a file with the output in tab as separated format, where the filename ends on.xls making it easily imported into spreadsheet programs. This file always contains the predicted peptides in the order they appeared in the input file. The output data for each peptide will be displayed on a single line with predictions for each of the selected alleles in different columns (Figure 3).
Figure 3.
Downloaded output sheet opened in Microsoft® Excel and adjusted with of column. The output was generated using input in Figure 1 and selecting the alleles HLA-A0201 and HLA-A0301. 10mer peptide predictions were chosen.
FINAL REMARKS
This server is developed to aid research and limit the resources needed for rational and effective CTL epitope discovery and will be continuously updated as new data become available. All comments and suggestions for usability improvements are most welcome.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was funded by European Commission (LSHB-CT-2003-503231, LSHB-CT-2004-012175) and National Institutes of Health (HHSNN26600400006C, HHSN266200400025C, HHSN266200400083C).
Conflict of interest statement. None declared.
REFERENCES
- 1.Lundegaard C, Lund O, Kesmir C, Brunak S, Nielsen M. Modeling the adaptive immune system: predictions and simulations. Bioinformatics. 2007;23:3265–3275. doi: 10.1093/bioinformatics/btm471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nielsen M, Lundegaard C, Worning P, Hvid CS, Lamberth K, Buus S, Brunak S, Lund O. Improved prediction of MHC class I and class II epitopes using a novel Gibbs sampling approach. Bioinformatics. 2004;20:1388–1397. doi: 10.1093/bioinformatics/bth100. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen M, Lundegaard C, Worning P, Lauemoller SL, Lamberth K, Buus S, Brunak S, Lund O. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12:1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peters B, Bui HH, Frankild S, Nielson M, Lundegaard C, Kostem E, Basch D, Lamberth K, Harndahl M, Fleri W, et al. A community resource benchmarking predictions of peptide binding to MHC-I molecules. PLoS Comput. Biol. 2006;2:e65. doi: 10.1371/journal.pcbi.0020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sylvester-Hvid C, Nielsen M, Lamberth K, Roder G, Justesen S, Lundegaard C, Worning P, Thomadsen H, Lund O, Brunak S, et al. SARS CTL vaccine candidates; HLA supertype-, genome-wide scanning and biochemical validation. Tissue Antigens. 2004;63:395–400. doi: 10.1111/j.0001-2815.2004.00221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sette A, Fleri W, Peters B, Sathiamurthy M, Bui HH, Wilson S. A roadmap for the immunomics of category A-C pathogens. Immunity. 2005;22:155–161. doi: 10.1016/j.immuni.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 8.Lundegaard C, Lund O, Nielsen M. Accurate approximation method for prediction of class I MHC af-finities for peptides of length 8, 10 and 11 using prediction tools trained on 9mers. Bioinformatics. 2008 doi: 10.1093/bioinformatics/btn128. in press, doi:10.1093/bioinformatics/btn128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.