Abstract
The regulatory sequence analysis tools (RSAT, http://rsat.ulb.ac.be/rsat/) is a software suite that integrates a wide collection of modular tools for the detection of cis-regulatory elements in genome sequences. The suite includes programs for sequence retrieval, pattern discovery, phylogenetic footprint detection, pattern matching, genome scanning and feature map drawing. Random controls can be performed with random gene selections or by generating random sequences according to a variety of background models (Bernoulli, Markov). Beyond the original word-based pattern-discovery tools (oligo-analysis and dyad-analysis), we recently added a battery of tools for matrix-based detection of cis-acting elements, with some original features (adaptive background models, Markov-chain estimation of P-values) that do not exist in other matrix-based scanning tools. The web server offers an intuitive interface, where each program can be accessed either separately or connected to the other tools. In addition, the tools are now available as web services, enabling their integration in programmatic workflows. Genomes are regularly updated from various genome repositories (NCBI and EnsEMBL) and 682 organisms are currently supported. Since 1998, the tools have been used by several hundreds of researchers from all over the world. Several predictions made with RSAT were validated experimentally and published.
INTRODUCTION
Noncoding DNA sequences play an essential role in all biological systems, by ensuring the spatial and temporal regulation of gene transcription. The interactions between transcription factor (TF) proteins and their target genes rely on the recognition of very short DNA signals, the cis-regulatory elements.
The regulatory sequence analysis tools (RSAT) offer a collection of specialized software applications for the detection of cis-acting regulatory elements in genomic sequences. The website supports various approaches to analyze noncoding sequences, including a variety of pattern discovery and pattern-matching programs. Pattern discovery (also called ab initio motif detection) takes as input a set of sequences, and detects exceptional motifs that are considered as putative regulatory signals. Pattern matching takes as input a set of sequences and a set of motifs (which may be obtained either from prior knowledge or by running a pattern-discovery program), and searches for instances of the motif in the sequences. These instances are considered as putative transcription factor-binding sites.
The web server has been running without interruption since May 1998. At that time, it was restricted to the yeast genome. More than 600 genomes are currently supported, and the data is regularly updated from various genome repositories (NCBI and EnsEMBL). In a previous description of the tools (1), the server was centered on the string-based pattern-discovery algorithms oligo-analysis (2) and dyad-analysis (3). RSAT have been recently upgraded by the inclusion of new tools for scanning sequences with position-specific scoring matrices (PSSMs), and for the detection of conserved elements in promoters of orthologous genes (phylogenetic footprints). A wide variety of genome- and taxon-specific background models are available, which provide the essential statistical background to assess the significance of the predicted motifs (pattern discovery) and sites (matrix-based pattern matching). In addition, the web interface has been recently redesigned to improve the navigation and offer a better accessibility to the programs.
We present hereafter a summary of the supported tools, with some examples of results obtained with the most recent applications.
TASKS AND PROGRAMS
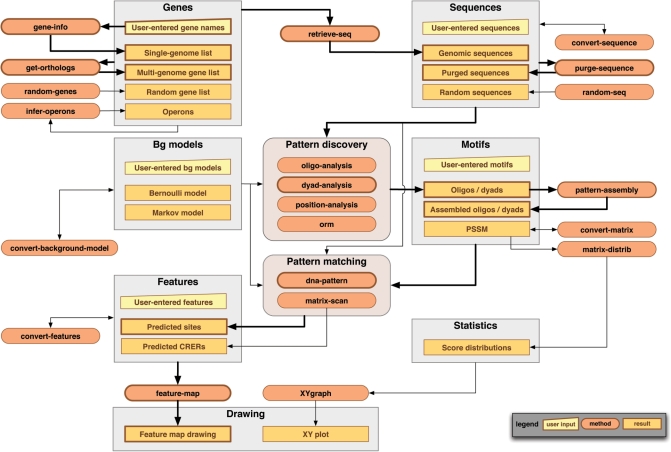
The procedures currently supported by RSAT are summarized in Table 1. Programs can be linked to build workflows as illustrated in Figure 1 or used separately according to each user's needs. We provide below a short description of the main program functionalities, with a specific emphasis on the tools that were not described in the previous publications about the RSAT web server (1,4).
Table 1.
Short description of the programs supported on RSAT web sites
| Task | Program name | Input | Output | Description |
|---|---|---|---|---|
| Genomes and genes | supported-organisms | Organism names | Returns the list of organisms supported on this site of rsa-tools | |
| gene-info | Gene names | Genes | Selects genes whose identifier, name or description matches a list of query strings. Partial matches are supported. | |
| infer-operons | Gene names | Operons + leader genes | Given one or more input genes, apply a simple distance-based rule to infer the operons to which those genes belong. Report the predicted operon leader gene and/or the complete operon. | |
| random-genes | Organism | Genes | Selects a random set of genes. | |
| get-orthologs | Given a gene or a list of genes from a query organism, and a reference taxon, this programs returns the orthologs of the query gene(s) in all the organisms belonging to the reference taxon | |||
| Sequences | retrieve-seq | Gene names | Sequences | Given a set of gene names, returns upstream, downstream or unspliced ORF sequences. The user defines the limits relative to the ORF start. Segments overlapping an upstream ORF can be excluded or included. |
| purge-sequence | Sequences | Sequences | Discards large repetitive fragments from a sequence set. Program developed by Stefan Kurtz. | |
| convert-seq | Sequences | Sequences | Interconversions between different sequence formats | |
| random-seq | Sequences | Generates random sequences. Different probabilistic models are proposed (equiprobable nucleotides, specific alphabet utilization and Markov chains). | ||
| Pattern discovery | oligo-analysis | Sequences | Exceptional oligos | Analyzes oligonucleotide occurrences in a set of sequences, and detects over- or under-represented oligonucleotides. Various background models and scoring statistics are supported. |
| dyad-analysis | Sequences | Exceptional dyads | Detects overrepresented dyads (spaced pairs of oligonucleotides) within a set of sequences. | |
| footprint-discovery | Sequences | Conserved dyads | Detects phylogenetic footprints by applying dyad-analysis in promoters of a set of orthologous genes. | |
| position-analysis | Sequences | Positionally biased oligos | Calculates the positional distribution of oligonucleotides in a set of sequences, and detects those which significantly deviate from a homogeneous distribution | |
| orm | Sequences | Locally over/under-represented oligos/dyads | Computes oligomer/dyad frequencies in a set of sequences, and detects locally over/underrepresented oligomers | |
| pattern-assembly | Oligos/dyads | Alignment | Aligns a set of strongly overlapping patterns (oligos or dyads). | |
| compare-patterns | String-based patterns (IUPAC) | Matches between patterns + related statistics | Counts matching residues between pairs of sequences/patterns from two sets, and assess the statistical significance of the matches. Patterns can be described using the IUPAC code for ambiguous nucleotides. Spaced patterns (dyads) are also supported. | |
| consensus | Sequences | PSSM | Detects shared motifs in unaligned sequences on the basis of a greedy algorithm. Developed by Jerry Hertz. | |
| gibbs | Sequences | PSSM | Detects shared motifs in unaligned sequences on the basis of a Gibbs sampling strategy. Developed by Andrew Neuwald. | |
| Pattern matching | dna-pattern | Sequences + multiple patterns (string description) | Matching positions in input sequences | String-based pattern matching program specialized for DNA sequences. IUPAC code for partially specified nucleotides is supported, as well as regular expressions. Several patterns can be searched simultaneously in several sequences, allowing a fast detection |
| genome-scale-dna-pattern | Multiple patterns (string description) | Matching positions in all upstream sequences | Pattern matching with dna-pattern, applied to all genes (upstream or downstream sequences) of a selected organism | |
| matrix-scan | Sequences + multiple patterns (PSSM) | Matching positions in input sequences | Scans sequences with one or several PSSMs to identify instances of the corresponding motifs (putative sites). This program supports a variety of background models (Bernoulli, Markov chains of any order). | |
| patser | Sequences + one pattern (PSSM) | Matching positions in input sequences | Pattern matching program based on a position-specific scoring matrix description of the patterns. Developed by Jerry Hertz. | |
| genome-scale-patser | Single pattern (PSSM) | Matching positions in all upstream sequences | Pattern matching with patser, applied to all genes (upstream or downstream sequences) of a selected organism | |
| convert-background-model | Background model | Background model | Interconversions between formats of background models supported by different programs. | |
| convert-features | Features | Features | Interconversions between various formats of feature description. | |
| compare-features | Features | Features + statistics | Compares two or more sets of features. This program takes as input several feature files (two or more), and calculates the intersection, union and difference between features. It also computes contingency tables and comparison statistics. | |
| convert-matrix | Patterns (PSSM) | Patterns (PSSM) | Performs inter-conversions between various formats of PSSMs. The program also performs a statistical analysis of the original matrix to provide different position-specific scores (weight, frequencies, information content) | |
| matrix-distrib | Patterns (PSSM) | Theoretical score distribution | Computes the theoretical distribution of score probabilities of a given PSSM. Score probabilities can be computed according to Bernoulli as well as Markov-chain background models | |
| Drawing | feature-map | Matching positions | Drawing | Draws a map with the results of pattern matching programs. Several sequences can be represented in parallel, allowing visual comparison of matching positions. |
| XYgraph | Numbers | Drawing | Draws a 2D graph from a table of numerical data |
Note that additional programs are available as Web Services and/or with the stand-alone tools.
Figure 1.
Flow chart of the regulatory sequence analysis tools. Rounded boxes represent programs, rectangles data and results and trapezoid user input. Bold arrows highlight the succession of tools used by the tool footprint-discovery.
Genome and gene information
Genomes are imported and regularly updated from various sources, mainly NCBI (for microbial genomes) and EnsEMBL (for higher organisms). In January 2008, 682 genomes were supported, including 578 bacteria, 49 archaea, 36 fungi, 13 metazoa, 2 alveolata and 1 plant. Genes can be specified according to their systematic identifiers, usual names or synonyms (as long as those are annotated in the source databases).
We recently added support for comparative genomics. The tool get-orthologs takes as input one or several query genes, and returns the list of genes with similar products in a given taxon. Pairwise similarities between peptidic sequences are precomputed using the gapped version of BLAST (5) and stored in RSAT genome repository. By default, the program returns the bidirectional best hits (BBH), which can be considered as putative orthologs. The BBH criterion can however be relaxed to collect paralogs as well. Alternatively, more stringent thresholds can be imposed on any statistics (bits, E-value, percent identity, etc.) returned by BLAST in order to impose restrictions on the reported similarities. The result of get-orthologs is a multi-genome list of genes, which can further be used as input by retrieve-seq.
For bacterial genomes, the program infer-operons permits to predict operons on the basis of a simple distance-based method (the distance can be specified by the user), and returns the composition of those predicted operons, together with their putative leader genes.
Sequence retrieval
The tool retrieve-seq allows retrieving noncoding sequences located upstream or downstream of query genes. By default, sequences are retrieved from the start (upstream) and stop (downstream) codons. For some organisms, the NCBI and EnsEMBL annotations include mRNAs start and end locations, which can then be used as references. Sequence lengths can either be specified as a fixed value, or be determined in a gene-specific way, depending on the distance to the neighbor gene. The program retrieve-seq has also been adapted to accept multi-genome queries, specified as a two-column input (the first column indicates the gene ID, the second column the organism name), such as the get-orthologs result file.
Sequences can be purged with the program purge-sequence, in order to mask redundant fragments. This program is a wrapper around the programs vmatch and mkvtree developed by Stefan Kurtz (6,7). Sequence purging is important for pattern discovery, since repeated copies of sequences introduce biases in the over- or under-representation statistics. In contrast, pattern matching is generally done on nonpurged sequences, since one wants to locate all instances of the searched motif.
Background models
The choice of the background model is a crucial parameter for both pattern discovery and pattern matching. Background models can be estimated either from the input sequences or from reference data sets. For each supported organism, RSAT provides a collection of precomputed background models for oligonucleotides (length 1–8 nt) as well as for dyads (monad length from 1 to 3 nt, spacing from 0 to 20 nt). These models were estimated on the basis of complete sets of upstream sequences. We recently added taxon-wide background models for the analysis of multi-genome data sets (8). Background models can also be imported from external programs, with the utility convert-background-model (Table 2).
Table 2.
Supported inter-conversions between formats
| Data type | Program name | Supported input formats | Supported output formats |
|---|---|---|---|
| Sequences | convert-seq | EMBL, fasta, multi, raw, tab, wconsensus | fasta, ig, multi, raw, tab, wconsensus |
| Features | convert-features | dna-pattern, feature-map, gff, gff3 | dna-pattern, feature-map, gff, gff3, fasta |
| PSSM | convert-matrix | AlignAce, pattern-assembly, cluster-buster, clustal, consensus, feature-map, gibbs, meme, MotifSampler, tab, TRANSFAC | consensus, patser, tab, TRANSFAC, SeqLogo |
| Background models | convert-background-model | oligo-analysis, MotifSampler, meme, dyad-analysis | transition table, oligo-analysis, patser, MotifSampler |
Pattern discovery
Since its origin, the RSAT project was centered on specialized algorithms for the discovery of cis-regulatory motifs from promoters of coregulated genes. Our first pattern-discovery algorithm, oligo-analysis, is based on the detection of overrepresented oligomers in nucleic or protein sequences (2). This program is time and memory efficient, and can be applied to genome-scale sequence sets (9). The approach was later extended to the detection of overrepresented spaced pairs, with the program dyad-analysis, which permits to detect spaced motifs such as those bound by fungal zinc cluster proteins (3) or bacterial helix–turn–helix factors (8,10). Relevant biological signals can also be detected on the basis of some positional specificity. The program position-analysis (9) allows the detection of biologically relevant signals based on a nonflat positional distribution. A new program, orm, combines positional information and analysis of over/underrepresentation, to detect motifs showing an exceptional frequency in restricted positional windows. The web server also integrates two pattern-discovery programs developed by third parties: consensus (11) and gibbs (12).
Phylogenetic footprint discovery
The pattern-discovery methods listed above were initially developed to predict motifs from a set of coregulated genes in a single organism. The increasing number of sequenced genomes now allows to apply pattern discovery in an ‘orthogonal’ way: starting from a single query gene in an organism of interest, collect its orthologs in a taxon of reference (e.g. all fungi), and detect overrepresented motifs in the promoters of these orthologs. This comparative genomic approach particularly gives good results with microbial genomes (8), because their promoter regions are generally short, and the number of sequenced genomes is now sufficient to obtain a reasonable signal-to-noise ratio. The program footprint-discovery runs a predefined workflow performing the required steps to discover overrepresented elements in promoters of the orthologs of one or several query genes.
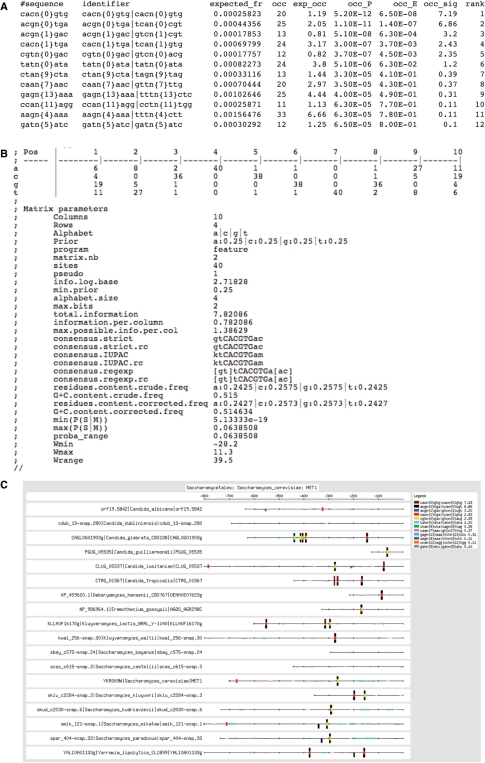
Figure 2 shows the result of footprints discovered in promoters of the orthologs of the gene MET1 in Saccharomycetales (Saccharomyces cerevisiae was used as query organism). Among the 43 680 possible dyads, 12 are significantly overrepresented in this set of promoters (Figure 2A). The feature map shows a strong overlap between instances of these dyads (Figure 2C), suggesting that they reveal alternative fragments of the same motif (3,8).
Figure 2.
Example of result from footprint-discovery. (A) overrepresented dyads detected in promoters of orthologs of the yeast gene MET1. (B) PSSM obtained by assembling the most significant dyads and using them as seeds to scan the input sequences. (C) Feature map of the significant dyads. The clumps of overlapping boxes are indicative of good predictions for binding sites.
A new feature of RSAT is that the string-based motifs resulting from dyad-analysis (or from oligo-analysis) can now be converted into PSSMs with the program matrix-from-patterns. This conversion relies on a three-step process: (i) a significance matrix is built from the assembled dyads (or oligonucleotides), by assigning to each cell of the matrix, the score of the most significant dyad containing the corresponding residue (row) at the corresponding position (column) of the aligned dyads; (ii) this significance matrix is used to scan input sequences for putative binding sites and (iii) putative binding sites are then aligned to form a count matrix. RSAT supports various formats for PSSMs (Table 2). In the tab-delimited format displayed in Figure 2B, the count matrix is documented by several statistical parameters (total information content, information per column, maximal weight, minimal weight, etc.).
Pattern matching
The program dna-pattern scans sequences with string-based patterns. This program supports various types of string-based patterns: single oligonucleotides, partly degenerated motifs (described with the IUPAC alphabet), spaced motifs or regular expressions. It can return a list of matches or a table showing the number of matches for each pattern (column) in each sequence (row).
The new program matrix-scan scans sequences with PSSMs, and scores each position according to the weight score previously defined by Jerry Hertz and Garry Stormo for their program patser (11,13,14), as well as the relative weight defined by Gert Thijs for MotifLocator (15). A particular strength of matrix-scan is its variety of supported background models, based on residue frequencies (Bernoulli) or higher-order dependencies between adjacent residues (Markov chains). Model estimation relies either on genome-wide reference sets (see ‘Background models’ section), or on the input sequence set.
RSAT matrix-based programs also support the computation of a P-value for each site, using either a Bernoulli or a Markov-chain model. The complete theoretical distribution of scores can be computed with matrix-distrib, in order to estimate the expected rate of false positives for each possible weight score.
In addition, matrix-scan allows to predict cis-regulatory modules by detecting genome segments enriched in PSSM matches (CRER, for cis-regulatory element enriched region). A P-value is associated to each CRER, using the binomial distribution of probability (16).
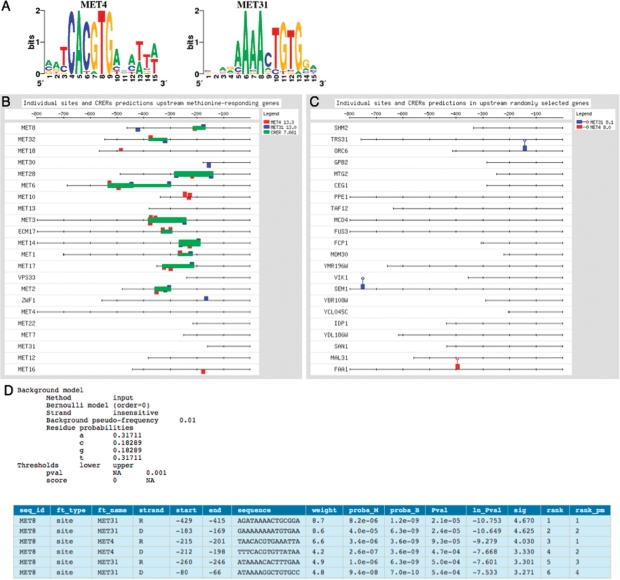
Figure 3 shows a typical result of a pattern-matching analysis conducted in RSAT. Upstream sequences of methionine-responding genes from Saccharomyces cerevisiae were scanned by matrix-scan with PSSMs describing the binding motifs of the transcription factors Met4p and Met31p (17) (Figure 3A). The predicted sites and CRERs (Figure 3D) were then sent to feature-map for graphical display. Figure 3B presents both the individual sites and CRER predictions. The random controls are shown in Figure 3C. Predicted sites found clustered in CRERs are likely to be putative sites for the transcription factors Met4p and Met31p. Consistently, matrix-scan predicts a high density of sites and CRERs upstream of the methionine-responding genes, whereas only three sites and no CRERs are predicted in the random controls. The latter predictions are probably false positives.
Figure 3.
Example of matrix-scan result obtained by scanning yeast upstream sequences with matrices representing binding motifs for the transcription factors Met4p and Met31p. (A) Sequence logos representing binding motifs of the Met4p and Met31p transcription factors. (B) Feature map of the predicted sites and CRERs in upstream sequences of 26 yeast genes involved in methionine metabolism. (C) Random control: feature map of the predicted sites and CRERs detected in upstream sequences of 26 yeast genes selected at random. (D) Fragment of a matrix-scan result table reporting putative sites.
Random controls
Random controls provide a powerful way to test the validity of the statistical models, by allowing to assess the rate of false predictions (false positives) returned by the program. One type of negative control consists in analyzing artificial sequences, generated at random according to some probabilistic model. The program random-seq generates random sequences according to any of the background models supported on RSAT.
Such random sequences with controllable properties are convenient to check the theoretical rate of false positives returned by a program (P-value, E-value), but they might fail to reflect the behavior of the same program on real biological sequences. Indeed, some biological sequences are too complex to be modeled by a simple Markov chain. A more realistic control can be achieved with random-genes. This program selects at random one or several gene sets, whose sequences can then be submitted to the same analysis workflows as those applied to clusters of coexpressed genes. In principle, a good predictive program should return significant results with coexpressed genes, and no result with randomly selected genes.
Drawing facilities
The web server includes two drawing tools: (i) feature-map generates graphical representations of features on sequences (e.g. predicted and/or annotated TF binding sites on promoter sequences) (e.g. Figures 2C, 3B and C); (ii) XYgraph generates XY plots from an input tab-delimited file.
Compatibility with other programs
A series of file converters ensures compatibility between RSAT and various formats produced by external programs: sequence files, feature files, background models, PSSMs (see Table 2 for currently supported input/output formats).
PROGRAMMATIC ACCESS TO RSAT THROUGH A WEB SERVICES INTERFACE
RSAT is also available as web services implemented using the standards SOAP (http://www.w3.org/TR/soap) and WSDL (http://www.w3.org/TR/wsdl). This type of access combines the advantages of the web server (no need for a local installation of programs and genomes) with those of stand-alone applications (possibility to automate the analytic flows and to iterate on multiple data sets). Users with basic skills in programming (notions of Perl, Python or Java) can easily write custom workflows that combine several tools exposed as web services. Such client programs can be written in any SOAP-supported language. In addition, workflows can be designed without any programming, using the graphical user interface of the program Taverna (18,19).
A typical web services session runs as follows: the client program starts by opening a connection to the remote RSAT server, then uploads user-specified data sets and sends a request to run a series of analyses with user-specified parameters. After completion of the analysis, the server sends the results back to the client. Furthermore, a client program can combine in a single workflow the tools available in RSAT and other bioinformatics resources exposed as web services.
A detailed documentation of the methods and parameters is provided on the web server (http://rsat.scmbb.ulb.ac.be/rsat/web_services/RSATWS_documentation.xml). Sample clients are available (http://rsat.scmbb.ulb.ac.be/rsat/web_services/RSATWS_clients.tar.gz) and the RSAT main tutorial includes a section explaining how to write client programs for web services (http://rsat.scmbb.ulb.ac.be/rsat/distrib/tutorial_shell_rsat.pdf).
DOCUMENTATION
When using bioinformatics programs, biologists are sometimes facing some difficulties to understand the meaning and impact of the parameters of a program or to interpret its results. Since the earliest versions of RSAT, we placed a particular effort on documenting the programs at different levels: demos, manuals, online tutorials and protocols. Each form of the web server includes one or several DEMO buttons, which automatically fill the form with typical data sets and parameters. The manual pages provide a comprehensive description of the options. Online tutorials guide new users through a step-by-step exploration of the tool functionalities, providing clues on the interpretation of the results, and warning them about critical issues and classical traps. We also published two protocols describing the utilization of the main tools (20,21).
SUMMARY AND PERSPECTIVES
As far as we know, RSAT is the most comprehensive existing resource for the analysis of regulatory sequences, at both levels of the diversity of tools and genome coverage.
Alternative web servers offering related facilities are usually restricted to a single pattern-discovery algorithm combined with some postprocessing companion utilities (pattern matching and pattern comparisons). For example, the BioProspector server (http://seqmotifs.stanford.edu/) combines a Gibbs-sampling pattern-discovery tool (22), with further adaptations to analyze phylogenetic footprints (CompareProspector) or chip-on-chip data (MDscan), respectively. The MEME server (23) combines an expectation–maximization pattern-discovery algorithm (24) with a matrix-based pattern-matching tool. Many web servers are also focused on a narrow range of species. For example, oPOSSUM supports human, worm and yeast (25,26). The eCis-analyst is specialized in the prediction of cis-regulatory modules in Drosophila melanogaster and D. pseudoobscura (27,28). A wider collection of tools is offered on the Zlab Gene Regulation Tools (http://zlab.bu.edu/zlab/gene.shtml), including cis-regulatory module detection with Cluster-Buster (29) and search for overrepresentation of PSSM hits with clover (30), rover and MotifViz (31).
The TOUCAN workbench (32,33) is a stand-alone application that combines sequence retrieval (from EnsEMBL), repeat masking, pattern discovery with MotifSampler (15), pattern matching, cis-regulatory module prediction and feature map drawing. TOUCAN can also be queried through a web services interface, and is able to access other remote resources. Actually, TOUCAN and RSAT can easily be interfaced via their respective web services interfaces. The last version of TOUCAN includes a remote utilization of oligo-analysis. Reciprocally, the demo workflows on the RSAT web server include some example of multi-program pattern discovery combining oligo-analysis (RSAT), dyad-analysis (RSAT) and MotifSampler (TOUCAN).
In the near future, our efforts will focus on increasing the inter-operability with other databases and web tools, by developing programmatic workflows using web services interfaces. The biggest challenge will undoubtedly be to cope with the ever-increasing pace of sequenced genomes, and to take advantage of these new resources to develop powerful methods for the analysis of regulatory sequences in higher organisms.
AVAILABILITY
The main server is located in Belgium (http://rsat.scmbb.ulb.ac.be/rsat/). Mirror servers are available in Mexico (http://embnet.ccg.unam.mx/rsa-tools/), Sweden (http://liv.bmc.uu.se/rsa-tools/), France (http://crfb.univ-mrs.fr/rsaTools/), Canada (http://rsat.ccb.sickkids.ca/) and South Africa (http://www.bi.up.ac.za/rsa-tools/). The RSAT web server is free and open to all users and there is no login requirement.
ACKNOWLEDGEMENTS
The RSAT project was originated at the Universidad Nacional Autonoma de Mexico, in the laboratory of Julio Collado-Vides, to whom J.v.H. is thankful for past and present collaboration. This work was supported by the Fonds pour la Formation à la Recherche dans l’Industrie et dans l’Agriculture, FRIA (PhD grants of R.J., S.B. and J.V.T.), the Vrije Universiteit Brussel (Geconcerteerde Onderzoeksactie 29) (M.T.-C., PhD grant). O.S. postdoc grant and E.V. research fellowship were funded by the BioSapiens Network of Excellence funded under the sixth Framework program of the European Communities (LSHG-CT-2003-503265). The postdoctoral grant of M.D. was funded by the Belgian Program on Interuniversity Attraction Poles, initiated by the Belgian Federal Science Policy Office, project P6/25 (BioMaGNet). Funding to pay the Open Access publication charges for this article was provided by Région Wallonne de Belgique (TransMaze project 415925).
Conflict of interest statement. None declared.
REFERENCES
- 1.van Helden J. Regulatory sequence analysis tools. Nucleic Acids Res. 2003;31:3593–3596. doi: 10.1093/nar/gkg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Helden J, Andre B, Collado-Vides J. Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol. 1998;281:827–842. doi: 10.1006/jmbi.1998.1947. [DOI] [PubMed] [Google Scholar]
- 3.van Helden J, Rios AF, Collado-Vides J. Discovering regulatory elements in non-coding sequences by analysis of spaced dyads. Nucleic Acids Res. 2000;28:1808–1818. doi: 10.1093/nar/28.8.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Helden J, Andre B, Collado-Vides J. A web site for the computational analysis of yeast regulatory sequences. Yeast. 2000;16:177–187. doi: 10.1002/(SICI)1097-0061(20000130)16:2<177::AID-YEA516>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 5.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurtz S, Schleiermacher C. REPuter: fast computation of maximal repeats in complete genomes. Bioinformatics. 1999;15:426–427. doi: 10.1093/bioinformatics/15.5.426. [DOI] [PubMed] [Google Scholar]
- 8.Janky R, van Helden J. Evaluation of phylogenetic footprint discovery for predicting bacterial cis-regulatory elements and revealing their evolution. BMC Bioinform. 2008;9:37. doi: 10.1186/1471-2105-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Helden J, del Olmo M, Perez-Ortin JE. Statistical analysis of yeast genomic downstream sequences reveals putative polyadenylation signals. Nucleic Acids Res. 2000;28:1000–1010. doi: 10.1093/nar/28.4.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benitez-Bellon E, Moreno-Hagelsieb G, Collado-Vides J. Evaluation of thresholds for the detection of binding sites for regulatory proteins in Escherichia coli K12 DNA. Genome Biol. 2002 doi: 10.1186/gb-2002-3-3-research0013. research0013.1–0013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hertz GZ, Hartzell G.W., III, Stormo GD. Identification of consensus patterns in unaligned DNA sequences known to be functionally related. Comput. Appl. Biosci. 1990;6:81–92. doi: 10.1093/bioinformatics/6.2.81. [DOI] [PubMed] [Google Scholar]
- 12.Neuwald AF, Liu JS, Lawrence CE. Gibbs motif sampling: detection of bacterial outer membrane protein repeats. Protein Sci. 1995;4:1618–1632. doi: 10.1002/pro.5560040820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 14.Stormo GD, Hartzell G.W., III Identifying protein-binding sites from unaligned DNA fragments. Proc. Natl Acad. Sci. USA. 1989;86:1183–1187. doi: 10.1073/pnas.86.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thijs G, Lescot M, Marchal K, Rombauts S, De Moor B, Rouze P, Moreau Y. A higher-order background model improves the detection of promoter regulatory elements by Gibbs sampling. Bioinformatics. 2001;17:1113–1122. doi: 10.1093/bioinformatics/17.12.1113. [DOI] [PubMed] [Google Scholar]
- 16.Aerts S, Van Loo P, Thijs G, Moreau Y, De Moor B. Computational detection of cis -regulatory modules. Bioinformatics. 2003;19(Suppl. 2):II5–II14. doi: 10.1093/bioinformatics/btg1052. [DOI] [PubMed] [Google Scholar]
- 17.Gonze D, Pinloche S, Gascuel O, van Helden J. Discrimination of yeast genes involved in methionine and phosphate metabolism on the basis of upstream motifs. Bioinformatics. 2005;21:3490–3500. doi: 10.1093/bioinformatics/bti558. [DOI] [PubMed] [Google Scholar]
- 18.Oinn T, Addis M, Ferris J, Marvin D, Senger M, Greenwood M, Carver T, Glover K, Pocock MR, Wipat A, et al. Taverna: a tool for the composition and enactment of bioinformatics workflows. Bioinformatics. 2004;20:3045–3054. doi: 10.1093/bioinformatics/bth361. [DOI] [PubMed] [Google Scholar]
- 19.Hull D, Wolstencroft K, Stevens R, Goble C, Pocock MR, Li P, Oinn T. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 2006;34:W729–W732. doi: 10.1093/nar/gkl320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sand O, van Helden J. Discovery of motifs in promoters of coregulated genes. Methods Mol. Biol. 2007;395:329–348. doi: 10.1007/978-1-59745-514-5_21. [DOI] [PubMed] [Google Scholar]
- 21.Janky R, van Helden J. Discovery of conserved motifs in promoters of orthologous genes in prokaryotes. Methods Mol. Biol. 2007;395:293–308. doi: 10.1007/978-1-59745-514-5_18. [DOI] [PubMed] [Google Scholar]
- 22.Liu X, Brutlag DL, Liu JS. BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput. 2001:127–138. [PubMed] [Google Scholar]
- 23.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bailey TL, Elkan C. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995;3:21–29. [PubMed] [Google Scholar]
- 25.Ho Sui SJ, Mortimer JR, Arenillas DJ, Brumm J, Walsh CJ, Kennedy BP, Wasserman WW. oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 2005;33:3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho Sui SJ, Fulton DL, Arenillas DJ, Kwon AT, Wasserman WW. oPOSSUM: integrated tools for analysis of regulatory motif over-representation. Nucleic Acids Res. 2007;35:W245–W252. doi: 10.1093/nar/gkm427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berman BP, Nibu Y, Pfeiffer BD, Tomancak P, Celniker SE, Levine M, Rubin GM, Eisen MB. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl Acad. Sci. USA. 2002;99:757–762. doi: 10.1073/pnas.231608898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berman BP, Pfeiffer BD, Laverty TR, Salzberg SL, Rubin GM, Eisen MB, Celniker SE. Computational identification of developmental enhancers: conservation and function of transcription factor binding-site clusters in Drosophila melanogaster and Drosophila pseudoobscura. Genome Biol. 2004;5:R61. doi: 10.1186/gb-2004-5-9-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frith MC, Li MC, Weng Z. Cluster-Buster: finding dense clusters of motifs in DNA sequences. Nucleic Acids Res. 2003;31:3666–3668. doi: 10.1093/nar/gkg540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frith MC, Fu Y, Yu L, Chen JF, Hansen U, Weng Z. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004;32:1372–1381. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu Y, Frith MC, Haverty PM, Weng Z. MotifViz: an analysis and visualization tool for motif discovery. Nucleic Acids Res. 2004;32:W420–W423. doi: 10.1093/nar/gkh426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aerts S, Thijs G, Coessens B, Staes M, Moreau Y, De Moor B. Toucan: deciphering the cis-regulatory logic of coregulated genes. Nucleic Acids Res. 2003;31:1753–1764. doi: 10.1093/nar/gkg268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aerts S, Van Loo P, Thijs G, Mayer H, de Martin R, Moreau Y, De Moor B. TOUCAN 2: the all-inclusive open source workbench for regulatory sequence analysis. Nucleic Acids Res. 2005;33:W393–W396. doi: 10.1093/nar/gki354. [DOI] [PMC free article] [PubMed] [Google Scholar]