Abstract
MADNet is a user-friendly data mining and visualization tool for rapid analysis of diverse high-throughput biological data such as microarray, phage display or even metagenome experiments. It presents biological information in the context of metabolic and signalling pathways, transcription factors and drug targets through minimal user input, consisting only of the file with the experimental data. These data are integrated with information stored in various biological databases such as NCBI nucleotide and protein databases, metabolic and signalling pathway databases (KEGG), transcription regulation (TRANSFAC©) and drug target database (DrugBank). MADNet is freely available for academic use at http://www.bioinfo.hr/madnet.
INTRODUCTION
Like all high-throughput analysis methods, microarray technology allows scientists to simultaneously study expression patterns of thousands of genes, or even entire genomes. In the last decade, most of the problems concerning experimental precision, accuracy and reproducibility of microarray experiments have been addressed through maturing technology and improvements in analysis algorithms (1,2). This resulted in a widespread presence of microarray-based experiments in both research and diagnostic laboratories (3). Still, the final step in analysis of microarray experiments—the biological interpretation of results, remains a lingering issue, especially when an increasing number of microarray users originate from wide areas of research fields, often quite distant from bioinformatics and statistics. With every gene descriptor leading to many different knowledge databases, researchers without sufficient operational knowledge of bioinformatics may find themselves in a forest of information too large to grasp or manipulate. Therefore, a lot of recent effort is directed into bringing the microarray results back to the experimentalists’ workbenches (4–9).
We have developed a data analysis and visualization tool for high-throughput experiments that should minimize necessary technical knowledge, as well as effort and time required to gain biological insight into a large amount of data. MADNet, the microarray database network eliminates the need for prior knowledge of the existent microarray data formats and gene nomenclature, allows simultaneous access to multiple biological databases, while providing an interactive and user-friendly interface with a strong emphasis on graphical data representation.
Moreover, MADNet is not only confined to microarray experiments, but also can be used to analyse expression information from different experimental techniques. MADNet can mine information from diverse origins such as nucleic acid and protein microarrays, SAGE, phage display, as well as any other experimental technique that measures differential change in expression of a series of genes or proteins. The only requirement for MADNet is an experimental data file containing columns with gene identification numbers, fold change descriptors and optional statistical significance. MADNet is highly processive and able to analyse large quantities of input data. Therefore, it can also be used to analyse whole metagenomes (e.g. in terms of abundance of functional gene categories or presence and absence of metabolic pathways), which is especially interesting in context of recent progress in environmental sample sequencing. Furthermore, authors are ready to promptly enter database of user's interest and make it available through the MADNet interface, in order to answer the personalized needs of different research communities.
Integrated databases
MADNet provides a systems biology approach to complex research problems in a user-friendly interface. The software tool integrates several types of biological information from existing databases: NCBI nucleotide and protein databases (10), metabolic and signalling pathway databases (KEGG—Kyoto Encyclopedia of Genes and Genomes) (11), transcription regulation (TRANSFAC©) (12) and a drug target database (DrugBank) (13).
These data are stored in a local MySQL (http://www.mysql.com) database and integrated with a PHP-based scripting system (http://www.php.net) into an interactive web-based graphical interface. Differential expression data from user input are colour coded and mapped onto metabolic and signalling pathways and displayed either as an interactive, user-clickable map or a list of metabolic and signalling pathways, ranked according to the statistical significance of expression magnitude. MADNet also provides the ability to investigate transcriptional cascades by summarizing and visualizing transcription factor gene regulation networks.
MADNet
Web server implementation
The web server user interface is available in the form of HTML pages, dynamically generated by a set of server-side PHP scripting programs, which access information stored in the MySQL database. Special attention has been paid to the ‘processivity’ of the analysis. MADNet can process tens of thousands of genes, in order of minutes with the bottleneck usually being the time taken to upload the input file to the server. Thereafter, user's query is stored in the form of a session, which significantly reduces the time required to perform repetitive calculations and increases the overall responsiveness of the server.
There is a possibility for a user to test MADNet capabilities by selecting a demonstration mode using a sample microarray file already uploaded on the server.
Input
First step in MADNet analysis is the upload of user's normalized differential expression data file, containing two columns with gene identifier and differential expression value (fold change) and, optionally, a third column with P-values (i.e. statistical significance of corresponding gene-expression values). Input file can be formatted as either tab-delimited or comma separated (CSV) text, while expression values can optionally be log-transformed.
MADNet will attempt to spare the user of unnecessary input and attempt to automatically recognize file format, header row, target organism, gene annotation type and format of expression values (i.e. whether the values are log-transformed or linear). This detection is performed by scanning the first 100 lines of the input file and attempting to match gene identifiers against the local database. The leftmost detected column with floating point numbers is assumed to contain expression data, and any successive columns with similar format are assumed to contain optional P-values. Due to data limitations and the unavoidable possibility of ambiguous detection, MADNet allows user interventions in the automatic detection process. Currently supported gene identifiers are: NCBI GenBank, NCBI RefSeq, NCBI GeneID, UniProt and ENSEMBL Gene. Affymetrix gene identifiers are not supported in the present release and user is encouraged to convert them to RefSeq identifiers.
Although MADNet can process files containing other columns with additional or irrelevant data, processing speed can be radically increased and time for file upload significantly reduced if such columns are removed manually (e.g. if there is a large number of extra columns and/or file size exceeds tens of megabytes).
Analysis and output
Upon the completion of automatic file format detection, user is presented with the summary and the possibility to confirm or modify the suggested parameters.
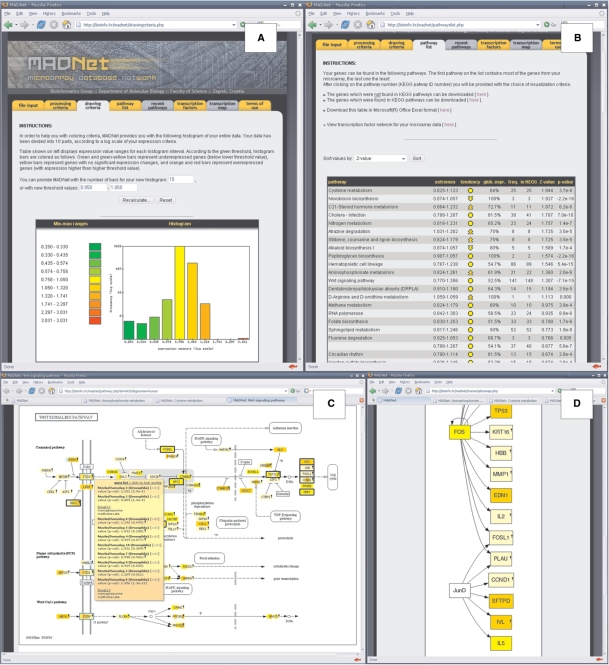
Based on two calculated threshold values, MADNet separates input data into three distinct categories: (i) the under-expressed genes (genes with the expression value less or equal than the lower threshold value); (ii) the over-expressed genes (genes with the expression value greater or equal than the higher threshold value) and (iii) genes with no significant change in the expression level (with expression value between the two thresholds). Default threshold values are automatically determined from the 2σ interval of binomial distribution of log-transformed expression values, and user is offered the possibility to manually adjust both thresholds independently. Threshold settings are accompanied with a histogram of expression values, colour coded to reflect expression value magnitude and thresholds—green-to-yellow ramp for under-expressed genes, yellow-to-red ramp for over-expressed genes and yellow for genes between the two thresholds (Figure 1A). These colours are consistently used throughout all subsequent visualization steps and can reliably be used as a visual cue while investigating expression value patterns on metabolic and signalling pathways.
Figure 1.
MADNet web server interface. (A) Overall expression histogram. After the processing of the input file, a fold change histogram is calculated to show general tendencies in the submitted data. User is also presented with the possibility to alter expression threshold values. (B) Pathway list, sorted according to the Z-score significance. Clicking on the pathway name opens a new browser tab or window with the pathway map. (C) Graphical representation of submitted data in the context of a metabolic or signalling pathway. Fold change values for genes found in a particular pathway are rendered in colour, according to the previously set threshold. Moving the mouse over the box with a gene name opens a pop-up window with detailed expression information and links to NCBI databases. (D) Dynamically generated transcription factor cascade map.
MADNet then sorts genes into corresponding metabolic and signalling pathways and offers a list of pathway names (Figure 1B) sorted according to the significance of regulation, computed from either the P-value of binomial distribution or Z-score (14). The list is supplemented with additional statistical indicators, such as extremes (minimum and maximum expression values for the respective pathway), tendency (median expression value, displayed as an arrow that demonstrates an overall regulation trend), frequency (gene count), ‘in KEGG’ (total number of genes found in the respective pathway in KEGG database), Z-score and P-value.
After clicking on a pathway name, an interactive graphical pathway map is opened in a new browser window (Figure 1C). The map contains annotations, cross-references with links to external databases and colour-coded expression values. Moving the mouse over a gene will produce a pop-up tooltip with a detailed list containing names of proteins on that location, expression value and P-values, known transcription factors and known drugs (in the case of the human genome) acting on the respective gene(s) or protein products, as well as a hyperlink to the NCBI Entrez Gene database offering additional detailed information.
User can go back to the pathway list and open in parallel as many pathways as desired. This is especially useful in cases when several different pathways are analysed simultaneously, or the same pathway is compared across different experiments. Furthermore, MADNet provides the user with the automatically generated complete reports of the analysed data, available in the Microsoft® Excel and tab-delimited text file formats, which is especially convenient for high-throughput analysis of a batch of experiments.
MADNet integrates the transcription factor database TRANSFAC© and uses it to identify and visualize known transcription cascades within user's data. Transcription factors are cross-referenced to the user submitted data in two ways: by (i) total number of regulated genes per transcription factor and (ii) average expression value for the regulated group of genes. From this point, user can follow three different investigation paths.
First, by selecting a particular transcription factor from the drop-down list and pressing the ‘Go to …’ button, user will be presented with a detailed break-down of genes and pathways found to be affected by the selected transcription factor. By selecting ‘view on pathways’ option, user is taken to a complete listing of metabolic and signalling pathways, containing only information pertinent to genes regulated with a selected transcription factor. The pathway map generated from the subset list will highlight only regulated genes for easier identification on complex pathways.
Second, in order to provide user with as detailed information as possible, clicking on either transcription factor or regulated gene will open the NCBI Entrez Gene web page.
Third, by selecting one or more transcription factors from the drop-down list of transcription factors and pressing ‘Submit’, user is presented with a dynamically generated transcription factor regulation network graph (Figure 1D). This network graph consists of nodes representing selected transcription factors and genes or other transcription factors they regulate, and directed lines, which indicate direction of regulation (i.e. lines pointing into a node connect that node to all nodes regulating it and lines pointing away from a node connect it to all nodes that are regulated by that node). Line pointing from one onto the same node represents self-regulation of that transcription factor.
CONCLUSIONS AND FUTURE WORK
MADNet, the microarray database network, is a versatile data mining and visualization web server for analysis of high-throughput experimental data. It integrates experimental results with the existing biological data in the context of metabolic and signalling pathways, transcription factors and drug targets and presents the results graphically, and in an intuitive, biology-centric way, with minimal technical requirements and without limits on the size of the experiment. Some of the novelties include DrugBank and TRANSFAC integration, the ability to process chips of unlimited length, several different statistical measurements of pathway alterations, and an extensible and modular system for including future database links and annotations.
Future work will include underlying database consolidation in terms of the gene identifiers, as well as adding new species mappings into the database structure. A major improvement foreseen in the following releases will include the dynamic rendering of MAPP formatted pathways with the possibility for analysis of user/submitted pathways. We also plan to include automatic recognition of all standard chip layouts and gene identifiers, further removing the number of steps needed to reach the visualization stage. Furthermore, MADNet can easily be adopted to visualize data in the context of functional categories, like Gene Ontology (GO) or Clusters of Orthologous Genes.
ACKNOWLEDGEMENTS
This work is funded by the EMBO Young Investigator Program (Installation grant 1431/2006 to K.V.) and Croatian MSES grant 119-0982913-1211. Funding to pay the Open Access publication charges for this article was provided by EMBO.
Conflict of interest statement. None declared.
REFERENCES
- 1.Slonim DK. From patterns to pathways: gene expression data analysis comes of age. Nat. Genet. 2002;32(Suppl):502–508. doi: 10.1038/ng1033. [DOI] [PubMed] [Google Scholar]
- 2.Yang YH, Speed T. Design issues for cDNA microarray experiments. Nat. Rev. Genet. 2002;3:579–588. doi: 10.1038/nrg863. [DOI] [PubMed] [Google Scholar]
- 3.Jaluria P, Konstantopoulos K, Betenbaugh M, Shiloach J. A perspective on microarrays: current applications, pitfalls, and potential uses. Microb. Cell Fact. 2007;6:4. doi: 10.1186/1475-2859-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouton CM, Pevsner J. DRAGON View: information visualization for annotated microarray data. Bioinformatics. 2002;18:323–324. doi: 10.1093/bioinformatics/18.2.323. [DOI] [PubMed] [Google Scholar]
- 5.Chung HJ, Park CH, Han MR, Lee S, Ohn JH, Kim J, Kim JH. ArrayXPath II: mapping and visualizing microarray gene-expression data with biomedical ontologies and integrated biological pathway resources using scalable vector graphics. Nucleic Acids Res. 2005;33:W621–W626. doi: 10.1093/nar/gki450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 7.Diehn M, Sherlock G, Binkley G, Jin H, Matese JC, Hernandez-Boussard T, Rees CA, Cherry JM, Botstein D, Brown PO, et al. SOURCE: a unified genomic resource of functional annotations, ontologies, and gene expression data. Nucleic Acids Res. 2003;31:219–223. doi: 10.1093/nar/gkg014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grosu P, Townsend JP, Hartl DL, Cavalieri D. Pathway processor: a tool for integrating whole-genome expression results into metabolic networks. Genome Res. 2002;12:1121–1126. doi: 10.1101/gr.226602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salomonis N, Hanspers K, Zambon AC, Vranizan K, Lawlor SC, Dahlquist KD, Doniger SW, Stuart J, Conklin BR, Pico AR. GenMAPP 2: new features and resources for pathway analysis. BMC Bioinform. 2007;8:217. doi: 10.1186/1471-2105-8-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aoki-Kinoshita KF, Kanehisa M. Gene annotation and pathway mapping in KEGG. Methods Mol. Biol. 2007;396:71–92. doi: 10.1007/978-1-59745-515-2_6. [DOI] [PubMed] [Google Scholar]
- 12.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]