Abstract
Many biological experiments and their subsequent analysis yield lists of genes or proteins that can potentially be important to the prognosis or diagnosis of certain diseases (e.g. cancer). Nowadays, information about the function of those genes or proteins may be already gathered in some databases, but it is essential to understand if some of the members of those lists have a function in common or if they belong to the same metabolic pathway. To help researchers filter those genes or proteins that have such information in common, we have developed PaLS (pathway and literature strainer, http://pals.bioinfo.cnio.es). PaLS takes a list or a set of lists of gene or protein identifiers and shows which ones share certain descriptors. Four publicly available databases have been used for this purpose: PubMed, which links genes with those articles that make reference to them; Gene Ontology, an annotated ontology of terms related to the cellular component, biological process or molecular function where those genes or proteins are involved; KEGG pathways and Reactome pathways. Those descriptors among these four sources of information that are shared by more members of the list (or lists) are highlighted by PaLS.
INTRODUCTION
Much of the software for the analysis of genomic data yields lists of genes or proteins relevant for a given disease. Information about common features of those genes on these lists may have been already stored in some of the databases publicly available. To help the user make sense of these lists, we have created PaLS (pathway and literature strainer), a new tool that filters out those descriptors that are more represented in the list.
Four different types of descriptors are considered in PaLS: Pubmed references (1), Gene Ontology (GO) terms (2), KEGG pathways (3), and Reactome pathways (4). They give an insight on the kinds of biological processes different genes and proteins are known to be involved in.
PaLS is particularly useful for the biological interpretation of results from studies of differential expression and, specially, gene selection in the context of classification and prediction with microarray data. In all of these cases, the final output are lists of ‘interesting genes’, either because there is evidence that those genes show differential expression among conditions, or because those genes can be used to classify patients (e.g. when attempting to predict good versus bad prognosis in cancer patients). Variable selection with microarray data (in general, in scenarios where the number of variables is much larger than the number of samples), however, can lead to many solutions that have similar prediction errors, but that share few common genes (5–9). In other words, repeated runs of the same algorithm (or of similar algorithms), often return different solutions: different lists of ‘interesting genes’ that share few, if any, genes. These different solutions, however, even if different in terms of the individual genes selected, are frequently equivalent in the sense that they lead to the same predictions for subjects and have similar estimated prediction error rates. This multiplicity of results (many lists of genes that share few genes) is not a problem when the only objective of our method is prediction, but it is problematic for the biological interpretability of the results (5). Which one of the solutions, or sets of interesting genes, should we choose? Moreover, choosing one set of genes without awareness of the multiple solutions can create a false perception that the selected set is distinct from the rest of the genes. Instead of focusing on the identity of the individual genes selected, PaLS allows us to try to discover the major biological themes (e.g. main biological pathways) that are shared among different solutions, even if the identity of the genes in each solution is different. For instance, it is straightforward to use PaLS on different cross-validation or bootstrap runs in a classification or prediction study. An example will be shown below.
There are other tools with a similar goal to PaLS: Genecodis (10) shows most cited GO terms, KEGG pathways, InterProf motifs and SwissProt keywords, but it has to be rerun for each database, making it cumbersome to use as an exploratory tool; GeneTools (11) and FatiGO (12) only consider GO terms and, thus, have a much more limited scope than PaLS; and DAVID Gene Functional Classification Tool (13) takes a list of identifiers and it clusters them into groups based on their common descriptors, while PaLS does it the other way around: it shows those descriptors that are common within each list of identifiers.
FUNCTIONALITY
The main input file for PaLS is a plain text file containing a list or several lists of gene or protein identifiers. Each list can have its own name, which has to appear at the top of the list, after the ‘#’ symbol. The types of identifiers accepted by the application are: Ensembl Gene IDs, UniGene cluster IDs, Gene names (HUGO), GenBank accessions, Clone IDs, Affymetrix IDs, EntrezGene IDs, RefSeq_RNAs, RefSeq_peptides, SwissProt names. Thus, all of the main types of identifiers in common use are covered by PaLS. If a given identifier from the input list is not found on the database it is removed from the analysis. Data for three different organisms is accepted: human, mouse and rat. Internally, using the database of pregenerated conversion of identifiers used by IDconverter (14), PaLS does all the necessary translation of identifiers using the paths displayed in http://pals.bioinfo.cnio.es/help/PaLS-schema.png.
PaLS has three different methods of filtering annotations:
Filter those descriptors that are referenced by more than a given percentage of identifiers, giving results for each list separately. This method of filtering is intended to be used as a way of discerning which list, among the lists that a predictive software can output, has some common previously published information that shows that those genes or proteins share a similar function.
Group all lists in one list (removing duplicates) and display those descriptors that are more referenced in this global list. This method allows the user to see commonalities even if they are not seen within each list.
Look for those descriptors that are referenced by more than a given threshold of identifiers in more than a given percentage of lists. This allows looking for commonalities present within and among sets of lists.
Threshold values for the percentage of appearance of each of the type of the descriptors are part of the input information needed, but they have a default value of 50%.
Specially for PubMed references, and due to the popular tendency of genomics articles citing thousands of genes, it is suggested to use lower thresholds to obtain results that may be more specific than general articles. For all types of descriptors, the most time consuming process is the first search; once this is completed, the user can change thresholds for each type of descriptor and filtering method, and obtain an answer in a short time.
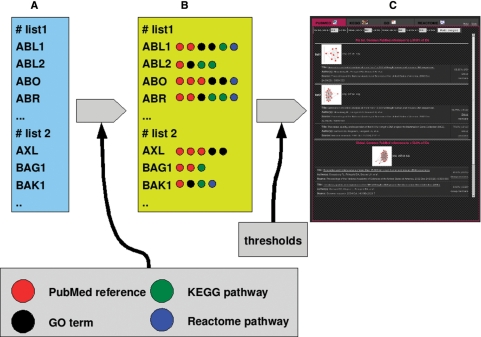
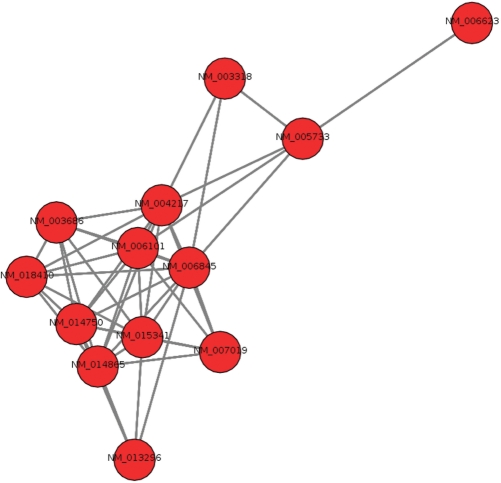
The output of PaLS are lists of those descriptors that fulfill the threshold criteria selected by the user, and the input identifiers related to each descriptor, linked to IDClight (14) to present the user with as much information as possible. This process is depicted in Figure 1. Also, for lists of less than 100 nodes, graph plots that describe the data structure of the lists are created. These plots (Figure 2), show all the genes or proteins in the list that have at least one descriptor as nodes. Two nodes (genes or proteins) are linked if they have descriptors in common; the more descriptors they share, the closer they appear.
Figure 1.
PaLS processing steps. Starting from a list or lists of protein or gene identifiers (A), PaLS looks for all their descriptors in the same database of ID conversions pregenerated for IDconverter (14) (B). Finally, it sorts those descriptors that appear more often in the lists, so the user can get an idea of the of the relevance of their lists (C). This example is done with a list of cancer-related genes available in the Help section of the web server.
Figure 2.
Example of a graph plot produced by PaLS (generated with the NetworkX package). The graph shows, for list from the 7th cross-validation run, those RefSeq_RNAs that are connected through common Gene Ontology terms. It can be seen how there is a central group of genes that share more terms (as they are closer to each other), and a gene, NM_006623, on the right side, that is only connected to another gene of the list.
We use GO as a controlled vocabulary. For a gene or protein, we reach GO through Ensembl which, in turn, depends on Uniprot. Thus, we make no attempt to incorporate the complete ontology (e.g. for a given gene not all the parent terms are included) nor its relationships. There are four reasons for our choice. First, we use the same approach as provided by a trusted, experienced source (Ensembl). Second, incorporating the complete ontology and/or the possible ontological relationships is not without additional problems, such as at what level of the ontology to search for commonalities or which of the five possible relationships to consider; by relying on the Ensembl mapping we avoid having to make this decisions ourselves. Third, since we provide links to AmiGO, it is easy for users to navigate the ontologies. Fourth, it would be possible to incorporate other sources of information like GO slims (http://www.geneontology.org/GO.slims.shtml), which are cut-down versions of GO which might be tailored for specific purposes; it should also be possible to add searches that attempt to incorporate more of the ontological information, or a more ‘semantically aware’ analysis. However, as explained above, we have tried to avoid imposing our own arbitrary decisions or our own semantical/ontological models and have preferred, instead, to rely on a standard source of GO annotations.
IMPLEMENTATION
PaLS is written in Python and uses AJAX to display the results, as well as NetworkX (https://networkx.lanl.gov), a Python package, to create the graph plots. The database server is MySQL and it is populated through a series of Perl and Python scripts (14). All the possible conversions are pregenerated in order to dramatically improve the response time of the application. The process of pregenerating these conversions is done every 2 months, following Ensembl's update schedule, using the latest version available of each of the databases used: Ensembl, UniGene, PubMed, KEGG, Reactome and Gene Ontology. For now, human, mouse and rat are the only organisms considered by PaLS; however, if needed or required by the community, the backbone of the same scripts that pregenerate the conversions for these three organisms could be used to add any of the tens of organisms for which there are data in Ensembl, provided that the corresponding UniGene databases are also available. All codes are available on request from the authors under the GPL license.
EXAMPLE
As an example, we have used our application SignS (15) with the data set from van't Veer et al. (16) as provided in Bair and Tibshirani (17). The original names contained a mixture of types of identifiers. For the example, we have mapped all possible identifiers to RefSeq_RNA. The results from SignS are available at http://signs.bioinfo.cnio.es/Examples/vantVeer-FCMS/results.html, and those from PaLS can be found at http://pals.bioinfo.cnio.es/Examples/vantVeer-FCMS/results.html.
At 50% threshold, GO terms in most lists refer to ‘nucleus’. At the 40% threshold, the term ‘cell cycle’ appears in several of the lists. As reported in the original van't Veer et al. paper (16), genes involved in cell cycle are upregulated in the poor prognosis signature, which agrees with the results from SignS and PaLS (the IDs correspond to a signature in SignS associated with decreased survival). If we drill down further, and set the threshold at 20%, we see that ‘mitosis’ appears in most of the lists; again, this is a functional annotation that the original publication describes as common in genes associated with poor prognosis. DNA repair (and, thus, GO term ‘signal transduction’, mentioned in the original reference) can be found in some lists at lower thresholds. If we examine PaLS results from Reactome, at the 20% threshold we see that ‘Cell cycle.Mitotic’ is abundant in most of the lists. Interestingly, one of the lists (the 6th cross-validation run) shows ‘E2F mediated regulation of DNA replication’, and van't Veer et al. mention cyclin E2. The fact that we need to use relatively low thresholds suggests, however, that there is no ‘dominant theme’ among the results: genes seem to belong to different groups in terms of their functional categories and pathways.
Figure 2 shows an example of the graph plots generated by PaLS, that display the connectivity structure of the input lists.
CONCLUSIONS AND FURTHER DEVELOPMENT
PaLS is a useful tool that helps researchers in their genomic analysis, improving the potential meaningfulness and biological interpretability of any list of genes or proteins that the analysis may yield. Given its different ways of sorting out those descriptors that are more relevant, PaLS can fulfill most of the filtering needs of the user, providing a list of useful descriptors that enrich the knowledge associated with a given list. It is important to note that this filtering is done without depending on any statistical model. Also, due to its modular characteristics, it can be easily improved with the addition of new types of descriptors.
Among some further worthwhile developments, either for public or private versions, PaLS could include queries to specific, tailored GO databases (such as GO slims) as well as filtered queries to PubMed, maybe using Natural Language Processing, or filtering papers according to some predetermined criteria that might be relevant in certain institutions or research environments.
ACKNOWLEDGEMENTS
We are grateful to Edward R. Morrissey for providing us with his knowledge of AJAX and his AJAX code implemented in Pomelo II (http://pomelo2.bioinfo.cnio.es). We also thank Alex Cañada for the web design. Finally, we want to thank many testers at CNIO and elsewhere for feedback on the applications and bug reports. Funding provided by Fundación de Investigación Médica Mutua Madrileña and Project TIC2003-09331-C02-02 of the Spanish Ministry of Education and Science (MEC). Funding to pay the Open Access publication charges for this article was provided by the EU project LSHG-CT-2004-512143.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2006;34 doi: 10.1093/nar/gkj158. (Database issue) D5–D12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig J, et al. Gene ontology: tool for the unification of biology. the gene ontology consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in kegg. Nucleic Acids Res. 2006;34 doi: 10.1093/nar/gkj102. (Database issue), D354–D357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joshi-Tope G, Gillespie M, Vastrik I, D'E;ustachio P, Schmidt E, deBono B, Jassal B, Gopinath GR, Wu GR, et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33 doi: 10.1093/nar/gki072. (Database issue), D428–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Somorjai RL, Dolenko B, Baumgartner R. Class prediction and discovery using gene microarray and proteomics mass spectroscopy data: curses, caveats, cautions. Bioinformatics. 2003;19:1484–1491. doi: 10.1093/bioinformatics/btg182. [DOI] [PubMed] [Google Scholar]
- 6.Pan KH, Lih CJ, Cohen SN. Effects of threshold choice on biological conclusions reached during analysis of gene expression by DNA microarrays. Proc. Natl. Acad. Sci. USA. 2005;102:8961–8965. doi: 10.1073/pnas.0502674102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Díaz-Uriarte R, Alvarez deAndrés S. Gene selection and classification of microarray data using random forest. BMC Bioinform. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ein-Dor L, Kela I, Getz G, Givol D, Domany E. Outcome signature genes in breast cancer: is there a unique set? Bioinformatics. 2005;21:171–178. doi: 10.1093/bioinformatics/bth469. [DOI] [PubMed] [Google Scholar]
- 9.Michiels S, Koscielny S, Hill C. Prediction of cancer outcome with microarrays: a multiple random validation strategy. Lancet. 2005;365:488–492. doi: 10.1016/S0140-6736(05)17866-0. [DOI] [PubMed] [Google Scholar]
- 10.Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. Genecodis: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biology. 2007;8:R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beisvag V, Junge FKR, Bergum H, Jolsum L, Lydersen S, Gunther C-C, Ramampiaro H, Langaas M, Sandvik AK, Laegreid A. Genetools – application for functional annotation and statistical hypothesis testing. BMC Bioinform. 2006;7:470. doi: 10.1186/1471-2105-7-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Shahrour F, Díaz-Uriarte R, Dopazo J. Fatigo: a web tool for finding significant associations of gene ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 13.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord GW, Roayaei J, Stephens R, Baseler MW, Lane CH, Lempicki RA. David gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene list. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alibés A, Yankilevich P, Cañada A, Díaz-Uriarte R. Idconverter and idclight: conversion and annotation of gene and protein ids. BMC Bioinform. 2007;8:9. doi: 10.1186/1471-2105-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Díaz-Uriarte R. Signs: a parallelized, open-source, freely available, web-based tool for gene selection and molecular signatures for survival and censored data. BMC Bioinform. 2008;9:30. doi: 10.1186/1471-2105-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van't;Veer LJ, Dai H, van deVijver MJ, He YD, Hart AA, Mao M, Peterse HL, van derKooy K, Marton MJ, Witteveen A, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 17.Bair E, Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol. 2004;2:0511–0522. doi: 10.1371/journal.pbio.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]