Abstract
We present an easy-to-use webserver that makes it possible to simultaneously use a number of state of the art methods for performing multiple alignment and secondary structure prediction for noncoding RNA sequences. This makes it possible to use the programs without having to download the code and get the programs to run. The results of all the programs are presented on a webpage and can easily be downloaded for further analysis. Additional measures are calculated for each program to make it easier to judge the individual predictions, and a consensus prediction taking all the programs into account is also calculated. This website is free and open to all users and there is no login requirement. The webserver can be found at: http://genome.ku.dk/resources/war.
INTRODUCTION
Over the past few years, different studies have shown how noncoding RNAs (ncRNA) are involved in gene expression, cell specialization, multi-drug resistance, splicing etc. in all living cells (1,2). For instance, only a small part of mammalian genomes encodes protein coding genes, but experiments have shown that a large fraction of the genomes is transcribed (3). Thus, there is potential for a large number of ncRNA transcripts, and there is computational evidence for thousands of structured RNAs in several vertebrate genomes (4–6).
This has given rise to an increased interest in ncRNAs, and since the structure of these molecules is tightly linked to their function, structure prediction methods have received much attention. Many different methods have been developed and they vary greatly in their approaches to the problem. Previously, methods for folding a single sequence by predicting the minimum free energy conformation were pursued [mfold (7), RNAfold (8)]. Today, comparative methods are the norm where a multiple alignment of a set of related RNA sequences is a part of the approach. Either the alignment is predicted alongside the consensus structure [i.e. the Sankoff-approach (9)] or an alignment is part of the input to the structure prediction [e.g. RNAalifold (10)].
The benefit of using comparative methods is that more information is available than for single sequence approaches. Many programs have been published over the past few years, and it can be difficult for a user to determine which one to use, to judge the different predictions, and sometimes even to run the programs. Since the performance of the different programs depend on many factors such as sequence length and identity, a specific program will not always perform best. Using an ensemble of programs therefore makes it easier to get a good idea of the correct result.
Here, we present an easy way to run a selection of methods and get a combined view of the predictions. The user simply inputs the sequences to be analyzed, and a selection of programs is automatically run on the dataset. The predictions are analyzed in various ways to make the output more informative for the user. The results are presented on a webpage, where one can easily download the different predictions and compare the relative performance of the individual programs. We also present a combined consensus prediction based on the results.
MATERIALS AND METHODS
The webserver for aligning structural RNAs (WAR) performs multiple alignment and secondary structure prediction on a dataset using a number of programs. The input to the server is the RNA sequences to be analyzed in Fasta format. The sequences can either be uploaded as a file or copy–pasted to a field on the webpage. The methods chosen are (in alphabetical order):
CMfinder (11): an algorithm based on expectation maximization using covariance models. Searches for RNA motifs combined with structure prediction based on both folding energy and covariation.
FoldalignM (12): based on the Sankoff approach, where the partition function (13) is used to calculate basepair probability matrices for each sequence. These matrices are then aligned using progressive alignment to produce a multiple alignment and predicted consensus structure. The approach is similar to PMcomp/PMmulti (14).
LaRA (15): a mathematical approach using Lagrangian transformed relaxation. The problem of optimizing alignment and structure is formulated as an integer programming problem, and a numerical optimization approach is used.
MASTR (16): a sampling approach using Markov chain Monte Carlo in a simulated annealing framework, where both structure and alignment is optimized by making small local changes. The score combines the log-likelihood of the alignment, a covariation term and the basepair probabilities.
RNAalifold (10) + ClustalW (17): ClustalW is one of the most widely used alignment programs. It performs progressive alignment using a simple guide tree. RNAalifold predicts the structure given in an alignment using both the free energy and a covariation measure to evaluate the basepairing regions.
RNAforester (18) + RNAcast (19): RNAforester performs multiple alignment based on an input set of sequences with secondary structures. The output is thus based on structural similarities. The input set is generated using RNAcast that predicts the common shape for all sequences and the energetically best structure for each sequence.
RNASampler (20): Possible stems are found for each sequence and the stems are then aligned by comparing all pairs of sequences. A conservation score considering both structure and sequence alignment measures the quality, and a structural alignment is built. Unpaired regions are aligned using ClustalW.
All the programs perform only global alignment, except for CMfinder which is capable of performing local multiple alignment. Optionally, one can use the webserver to perform local alignment. This is done by extracting the best scoring local motif predicted by CMfinder. Then all the selected programs in the webserver are run globally, as usual, on the local region selected by CMfinder.
All the programs are run using their default settings. If the user wants to try other parameter settings, we encourage the use of the webservers and/or source code of the individual programs. There are a few limitations on the use of WAR: the user must submit at least two sequences (note that CMfinder does not work with less than three sequences), no more than 50 sequences can be submitted in one job and a sequence can be no more than 250 nucleotides in length unless the local alignment box is checked; in that case, there is no length limit.
Postprocessing of the results
For each method, the result is presented and can be easily downloaded for further use. The multiple alignment is colored using the coloraln-script from the Vienna package (8) and is shown with a barplot visualizing the conservation for each column. The consensus structure is written on top of the alignment, and the predicted basepairs are color coded to highlight canonical basepairs (i.e. Watson–Crick interactions and GU-basepairs) and compensatory mutations.
The consensus sequence is shown along with the consensus structure as predicted by the program (both as dot-bracket and a Postscript-file). The consensus sequence is defined using the whole range of IUPAC ambiguity characters (21) and is similar to the most informative sequence (22).
To quantify the quality of the alignment, the average pairwise identity is calculated. When a reference alignment is not known, it is not a trivial task to measure the correctness of an alignment. This is one of the reasons why so many different alignment algorithms exist. The identity measure used in the WAR server is not as such a measure of correctness, but instead a measure of the quality of the alignment. For instance, the correct alignment of highly diverged sequences will by necessity have a lower overall identity than the correct alignment of closely related sequences. However, in the current setting, a number of different methods have all been used on the same dataset and one can get an idea of the quality by comparing the pairwise identity of the alignments. If one program obtains a comparatively low pairwise identity, the alignment is probably worse. The average pairwise identity is calculated by making all the pairwise comparisons between sequences in the multiple alignment and counting the number of aligned positions that are identical. The average fraction of identities is then reported for the whole alignment.
To estimate the thermodynamical stability of the predicted structure, the average free energy is used. The average free energy in itself cannot be used directly as a quality measure, but by comparing the average of the different predictions the relative performance of each can be assessed. For a given prediction, we map the consensus structure to each sequence in the alignment after removing the gaps. The sequence is then folded into this specified structure and the free energy calculated using RNAeval (8). The average free energy for the entire alignment given the predicted consensus structure is then reported.
To evaluate how well the sequence alignment supports the predicted structure, we calculate a covariation score for each basepair, given the alignment. The proposed structure will pair up columns in the alignment, and covariation measures the amount of evidence for a basepair. This is done by calculating how often a variation in one column leads to a variation in the other. We use the measure that proved to be best in a recent study comparing different covariation measures (23) and report the average covariation score for all basepairs. Note that this measure can be negative (due to a penalty term) and greater than 1.
The quality of the predicted structure is also measured as the average basepair probability. For each sequence, we calculate the basepair probability matrix using RNAfold (8,13). For a single sequence, we can then find the probability of each proposed basepair by simply looking in the matrix. For two pairing columns in the alignment, the probability of a basepair is then found as the average probability for that particular basepair in each sequence. The average probability for the whole structure is then reported.
All these results are reported on a single webpage that makes it easy to compare the different methods for both similarities and differences. Alignments and structures are easily downloaded in different formats.
A consensus prediction is made based on all the programs in the following way: T-Coffee (24) is a program that performs multiple alignment using a library of all both local and global pairwise alignments in the set. The library is extended by realigning to a third sequence and weights are calculated based on consistencies within the library. The multiple alignment is then performed progressively based on these weights.
This method can also predict a consensus multiple alignment by building the library from a number of multiple alignments instead. A single consensus multiple alignment is constructed by giving the alignments from all the programs as input to T-Coffee. The consensus structure is found by taking each ungapped sequence and mapping the predicted structure from each program onto it. If a basepair is predicted by at least 50% of the programs, it becomes a part of the consensus structure for that sequence. This gives us a consensus structure based on all the programs for each sequence in the alignment. Each sequence is then aligned to the T-Coffee alignment, making it possible to compare the consensus structure for each sequence, and the basepairs that are present in at least 50% of the sequences become part of the consensus structure for the whole alignment.
WEBSERVER
We illustrate the use of WAR in the following and especially show how the consensus prediction can be useful. For this purpose, we use a tRNA dataset consisting of 10 randomly chosen sequences, but the procedure is the same for any dataset.
If it is known, a reference alignment and structure can be uploaded along with the unaligned sequences. This makes it possible to compare the predictions to the correct answer and thus rank the methods. We use the following scores: the predicted alignment is compared to the reference alignment using the sum of pairs score (SPS), which is a sensitivity-like measure (25). It is based on the fraction of nucleotide pairs aligned in the prediction that are also present in the reference and yields a number between 0 and 1, where 1 is perfect prediction. The predicted structure is compared to the reference using Matthew's; correlation coefficient (MCC), which shows the balance between sensitivity (SEN, i.e. the fraction of correct basepairs that are recovered by the method) and positive predictive value (PPV, i.e. the fraction of predicted basepairs that are also in the reference). MCC lies between −1 and 1, where 1 is perfect prediction.
Using WAR is simple: on the input form, you have to specify the input sequences either in the box or as a file. You also have to input a valid email address to receive notification when the job is complete (note that the email is used only for this notification). There are also some optional settings: you can choose to perform local alignment, you can specify a reference alignment file if available, you can name the submission and you can choose only to run a selection of the programs.
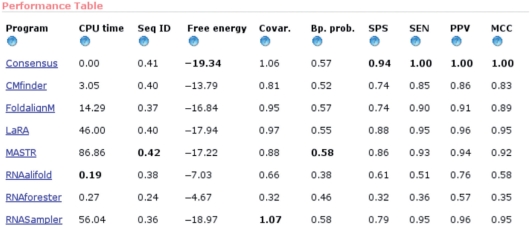
When the programs have finished, a table is shown that summarizes the predictions (Figure 1). If a reference alignment was given, the four rightmost columns summarize the performance (SPS, SEN, PPV and MCC). Otherwise, only the first six columns are shown (program, CPU time, average sequence identity, average free energy, covariation and average basepair probability). These measures are calculated as described earlier.
Figure 1.
The table showing the results from the different programs.
In the current example, LaRA and RNASampler gave the most correct structure predictions with MCC = 0.95. This corresponds with the two programs having the two lowest average free energies (−17.94 and −18.75, respectively) and the two highest average covariations (0.97 and 1.06, respectively). The average basepair probabilities are also high in both cases (0.55 and 0.58, respectively). The most correct alignment was also predicted by LaRA with SPS = 0.88. The measure of pairwise identity in the prediction is in this case 0.40, which is only the second largest. Notice, however, that the program with the highest pairwise identity also shows a very good SPS of 0.86.
When looking at the consensus prediction, it is evident that combining the structure predictions from all seven programs yields an improvement in both SEN, PPV and MCC to 1.0, which is better than the previous best prediction with MCC = 0.95. Also, the alignment is improved to SPS = 0.94, which is better than the single best prediction with SPS = 0.88. Looking at the other measures, the average free energy (−19.34) and covariation (1.06) are also best for the consensus, with the pairwise sequence identity (0.41) and basepair probability (0.57) being second best. In this case, using the consensus prediction based on the seven programs is clearly an improvement to using any single program.
Of course, that is not always the case, especially when only a few of the programs make reasonable predictions. Therefore, it is possible to update the consensus (see below) using only selected programs and not, as per default, all of them. For example, if some of the programs predict a very unstable consensus structure with a high free energy, it might be a good idea to update the consensus by removing those programs that perform poorly.
Clicking on any of the links in the first column shows a detailed description of the prediction from the chosen program. These pages are similar for all programs, only the consensus link differs. On the consensus page, the alignment is shown as a heat map at the top illustrating the confidence of the different parts of the alignment (from blue being low to red being high, see Figure 2). If necessary, one can choose which program to include in the consensus and update the alignment and structure. If some of the predictions are very different from the rest, the consensus might be improved by excluding these from the calculation.
Figure 2.
The top of the consensus result webpage showing the alignment as a heatmap.
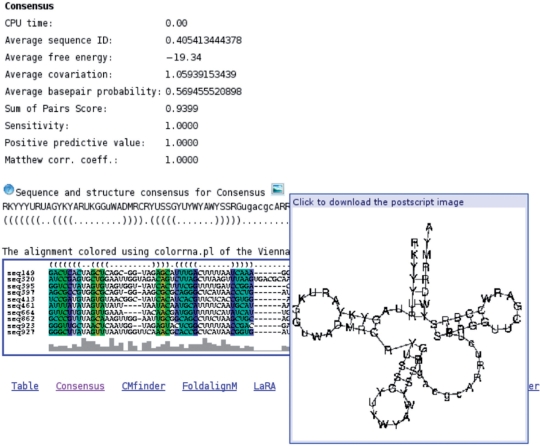
Further down the page is additional information, which is calculated for each program (Figure 3). The measures such as average pairwise identity and—if a reference was uploaded—MCC etc. are shown. Furthermore, the predicted consensus structure is shown with the calculated consensus sequence. By pointing with the mouse at the small image-icon, the structure is shown in a small pop-up window and by clicking it is downloaded as a Postscript-file. Below the structure a color-coded image of the alignment is shown, which can be downloaded as a gif-file or a Postscript-file. The raw output data from the program can be downloaded in various formats (Fasta, Clustal, Stockholm and col) in the top right corner of the page.
Figure 3.
The part of the consensus result page showing the predicted structure (with the pop-up window visible), the consensus sequence and the color-coded alignment. The consensus sequence is calculated from the actual nucleotide frequencies in each alignment column compared to the background frequencies to find overrepresented nucleotides. For instance, if only nucleotide ‘A’ is overrepresented, an ‘A’ will be added to the consensus. If, on the other hand, both ‘A’ and ‘C’ are overrepresented, an ‘M’ is used in the consensus. If more than half the sequences contain a gap in a column, a lower case letter is used. Otherwise, we use upper case. This part of the page is similar to the result pages for the other programs.
The consensus prediction is particularly useful when studying unknown structural RNAs. With no prior knowledge, it is hard to judge if getting a good prediction from a single program is reliable. But if all or several of the programs agree, and the heatmap shows a reliable consensus alignment and structure, you can have higher confidence in the results.
CONCLUSION
The WAR webserver makes it easy to use a number of methods for aligning and predicting the secondary structure for a set of structural RNAs. With all the focus on ncRNAs, this is a very useful tool for any researcher who wants to analyze sequences, but does not want to get the different programs to run him/herself. In time, WAR will be extended with new methods that perform well, and other combinations of alignment tools and structure prediction tools might be pursued.
What makes WAR especially useful is the postprocessing. The different measures calculated as part of the pipeline make it easier for the user to judge the quality of both the alignment and predicted structure, as well as compare the different methods. The calculated consensus alignment and structure is a valuable indicator of the quality of the predictions, especially for new, unknown sequences, where the user can get a good idea of how trustworthy the predictions are. Finally, WAR makes it easy to download the alignments and structures, both for the individual programs and the consensus, for further analysis.
It should be stressed again that the performance of the individual methods depend strongly on the actual dataset: the RNA family, the sequence length, the overall identity—all of this will affect the performance. The strength of the WAR server is the ease with which the different methods can be compared on different RNA datasets. In the example above, the tRNA dataset is fairly divergent and will thus be hard for methods that rely on a good, purely sequence-based alignment. A set of more closely related sequences will on the other hand be a challenge to programs that need a strong evolutionary signal from covariation. The goal of the above example is not to compare the individual methods but to show how the server works. On the website, the result of a different, less divergent dataset is available as an example.
ACKNOWLEDGEMENTS
We thank Jan Gorodkin, Stefan Seemann and Jakob S. Pedersen for their comments, and Paul Gardner for some rewarding conversations. Funding to pay the Open Access publication charges for this article was provided by a faculty PhD stipend.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bompfünewerer AF, Backofen R, Bernhart SH, Hertel J, Hofacker IL, Stadler PF, Will S. Variations on RNA folding and alignment: lessons from Benasque. J. Math. Biol. 2007;56:129–144. doi: 10.1007/s00285-007-0107-5. [DOI] [PubMed] [Google Scholar]
- 2.Bompfünewerer AF, Flamm C, Fried C, Fritzsch G, Hofacker IL, Lehmann J, Missal K, Mosig A, Müller B, Prohaska S, et al. Evolutionary patterns of non-coding RNAs. Theor. Biosci. 2005;123:301–369. doi: 10.1016/j.thbio.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 4.Washietl S, Hofacker IL, Lükasser M, Huttenhofer A, Stadler PF. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat. Biotechnol. 2005;23:1383–1390. doi: 10.1038/nbt1144. [DOI] [PubMed] [Google Scholar]
- 5.Pedersen JS, Bejerano G, Siepel A, Rosenbloom K, Lindblad-Toh K, Lander ES, Kent J, Miller W, Haussler D. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput. Biol. 2006;2:e33. doi: 10.1371/journal.pcbi.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torarinsson E, Sawera M, Havgaard JH, Fredholm M, Gorodkin J. Thousands of corresponding human and mouse genomic regions unalignable in primary sequence contain common RNA structure. Genome Res. 2006;16:885–889. doi: 10.1101/gr.5226606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sankoff D. Simultaneous solution of the RNA folding, alignment and protosequence problems. SIAM J. Appl. Math. 1985;45:810–825. [Google Scholar]
- 10.Hofacker IL, Fekete M, Stadler PF. Secondary structure prediction for aligned RNA sequences. J. Mol. Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 11.Yao Z, Weinberg Z, Ruzzo WL. CMfinder – a covariance model based RNA motif finding algorithm. Bioinformatics. 2006;22:445–452. doi: 10.1093/bioinformatics/btk008. [DOI] [PubMed] [Google Scholar]
- 12.Torarinsson E, Havgaard JH, Gorodkin J. Multiple structural alignment and clustering of RNA sequences. Bioinformatics. 2007;23:926–932. doi: 10.1093/bioinformatics/btm049. [DOI] [PubMed] [Google Scholar]
- 13.McCaskill JS. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990;29:1105–1119. doi: 10.1002/bip.360290621. [DOI] [PubMed] [Google Scholar]
- 14.Hofacker IL, Bernhart SHF, Stadler PF. Alignment of RNA base pairing probability matrices. Bioinformatics. 2004;20:2222–2227. doi: 10.1093/bioinformatics/bth229. [DOI] [PubMed] [Google Scholar]
- 15.Bauer M, Klau GW, Reinert K. Accurate multiple sequence-structure alignment of RNA sequences using combinatorial optimization. BMC Bioinform. 2007;8 doi: 10.1186/1471-2105-8-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindgreen S, Gardner PP, Krogh A. MASTR: multiple alignment and structure prediction of non-coding RNAs using simulated annealing. Bioinformatics. 2007;23:3304–3311. doi: 10.1093/bioinformatics/btm525. [DOI] [PubMed] [Google Scholar]
- 17.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Höchsmann M, Töller T, Giegerich R, Kurtz S. In Proceedings of the IEEE Bioinformatics Conference. Stanford, CA: Stanford University; 2003. Local similarity of RNA secondary structures; pp. 159–168. [PubMed] [Google Scholar]
- 19.Reeder J, Giegerich R. Consensus shapes: an alternative to the Sankoff algorithm for RNA consensus structure prediction. Bioinformatics. 2005;21:3516–3523. doi: 10.1093/bioinformatics/bti577. [DOI] [PubMed] [Google Scholar]
- 20.Xu X, Ji Y, Stormo GD. RNA Sampler: a new sampling based algorithm for common RNA secondary structure prediction and structural alignment. Bioinformatics. 2007;23:1883–1891. doi: 10.1093/bioinformatics/btm272. [DOI] [PubMed] [Google Scholar]
- 21.Cornish-Bowden A. Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984. Nucleic Acids Res. 1985;13:3021–3030. doi: 10.1093/nar/13.9.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freyhult E, Moulton V, Gardner PP. Predicting RNA structure using mutual information. Appl. Bioinform. 2005;4:53–59. doi: 10.2165/00822942-200504010-00006. [DOI] [PubMed] [Google Scholar]
- 23.Lindgreen S, Gardner PP, Krogh A. Measuring covariation in RNA alignments: physical realism improves information measures. Bioinformatics. 2006;22:2988–2995. doi: 10.1093/bioinformatics/btl514. [DOI] [PubMed] [Google Scholar]
- 24.Notredame C, Higgins D, Heringa J. T-Coffee: a novel method for multiple sequence alignments. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 25.Thompson JD, Plewniak F, Poch O. A comprehensive comparison of multiple sequence alignment programs. Nucleic Acids Res. 1999;27:2682–2690. doi: 10.1093/nar/27.13.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]