Abstract
The drug classification scheme of the World Health Organization (WHO) [Anatomical Therapeutic Chemical (ATC)-code] connects chemical classification and therapeutic approach. It is generally accepted that compounds with similar physicochemical properties exhibit similar biological activity. If this hypothesis holds true for drugs, then the ATC-code, the putative medical indication area and potentially the medical target should be predictable on the basis of structural similarity. We have validated that the prediction of the drug class is reliable for WHO-classified drugs. The reliability of the predicted medical effects of the compounds increases with a rising number of (physico-) chemical properties similar to a drug with known function. The web-server translates a user-defined molecule into a structural fingerprint that is compared to about 6300 drugs, which are enriched by 7300 links to molecular targets of the drugs, derived through text mining followed by manual curation. Links to the affected pathways are provided. The similarity to the medical compounds is expressed by the Tanimoto coefficient that gives the structural similarity of two compounds. A similarity score higher than 0.85 results in correct ATC prediction for 81% of all cases. As the biological effect is well predictable, if the structural similarity is sufficient, the web-server allows prognoses about the medical indication area of novel compounds and to find new leads for known targets.
Availability: the system is freely accessible at http://bioinformatics.charite.de/superpred. SuperPred can be obtained via a Creative Commons Attribution Noncommercial-Share Alike 3.0 License.
INTRODUCTION
The accessibility of large compound databases has changed from exclusive inhouse databases of large pharmaceutical companies to publicly available sources (1). At this time several million different compounds can be obtained from different vendors (2). About 7000 drugs currently exist and there are about 480 validated targets that are addressed (3). There are estimations about the number of medical targets between 2200 and 3000 that favour interactions with drug-like chemical compounds (4). To map these medical targets onto medical indication areas a classification scheme is needed.
Currently, the most commonly used classification system for drugs is the Anatomical Therapeutic Chemical (ATC) classification system. This scheme is recommended by the World Health Organization (WHO) for all global drug utilization studies and categorizes drug substances at different levels according to application area, therapeutic properties, chemical and pharmacological properties (5).
A challenging aim is the mapping of the available compounds onto about 850 ATC-classes. The progress in understanding the mechanisms of action of a vast majority of drugs gives the opportunity to narrow down the gap between the medical indications and elucidation of drug effects at the molecular level. The relation between the structure of a compound and its biological activity was well investigated in some systematic analyses (6–8). It could be shown that a Tanimoto coefficient of >0.85 indicates that two molecules have similar activities (8). Based on this principle, it should be possible to predict medical indication areas for unclassified chemical compounds in case of sufficient structural similarity. A method based on the similar property principle (9) for predicting activity spectra of substances was described by Lagunin (10) and confirmed by several experiments (11,12). The PASS application is available at http://www.ibmc.msk.ru/PASS. Furthermore, new medical indication areas for approved drugs or drug candidates can be found by applying this rule. Indeed recently, much efforts have been put into drug repositioning (13,14). To discriminate between drugs and nondrugs, the use of property distributions and (physico-) chemical descriptors is already used successfully (15,16). The increased knowledge about drug-target-pathway relations and the integration of molecular similarity with property distribution allow improved structure–function prediction. Here, we present a publicly available web-server to predict medical indication areas based on properties and similarity of chemical compounds.
METHODS
Data set for the web-server
The web-server called SuperPred was created for recognition of as many drug classes as possible. For this reason, the number of medical compounds was enlarged to about 6300. The calculated fingerprints from the 2500 compounds of the SuperDrug database were used for a further structural screening against the SuperTarget database (17). In this way, 3800 additional compounds were detected that are structurally very similar to drugs and resulted in Tanimoto coefficients of at least 0.85. These putative drugs are most likely candidates for having the same mode of action, binding to the same target/enzyme and being assigned to the same medical indication as the WHO-classified drugs. In order to allow the examination of the drug effect on a molecular level, information about the target proteins was extracted from literature and was provided for half of the drugs (17).
Reduced data set for prediction evaluation
For the purpose of statistical evaluation of the prediction accuracy, a subset consisting of 1035 drugs was utilized. The members of the subset were chosen according to the following rules: First, every drug having more than one indication was removed; then each included ATC-group had to consist of at least 3 molecules and last, to eliminate outliers, the drugs within one ATC-group were not allowed to deviate more than 1.5-fold from the average Tanimoto score of the group. Furthermore, ATC-codes with very similar indications were organized into ATC-classes. For instance, ‘corticosteroids, moderately potent’ (ATC: D07AB), ‘corticosteroids, potent’ (ATC: D07AC), ‘corticosteroids, very potent’ (ATC: D07AD) and ‘corticosteroids, plain’ (ATC: S01BA) were combined to form the ATC-class ‘corticosteroids’ (see website and Supplemental material).
Prediction
The prediction was carried out by the combination of physicochemical property analyses and similarity searching. The prediction of the ATC-class was performed by the assignment of the compound to the ATC-class of the most similar drug and property distribution. The prediction accuracy was determined by the leave-one-out cross-validation method.
Physicochemical properties
Lipinski's Rule of Five (18) is a general accepted standard for oral applicable drugs. The rule describes molecular properties important for a drug's absorption, distribution, metabolism and excretion in the human body. It is stated that an orally active drug has not more than 5 hydrogen-bond donors, not more than 10 hydrogen-bond acceptors, a molecular weight below 500 g/mol and a logP less than 5. These properties and several more were calculated for each drug in SuperPred. The distributions of the properties’ values were saved in the database for each ATC-group and -class. In this way, the range of property values is comparable with the query.
Similarity searches
To calculate the similarity between two compounds, their structural fingerprints, generated by Chemistry Development ToolKit (CDK) (http://almost.cubic.uni-koeln.de/cdk/), were used. Structural fingerprints are bit-vectors encoding for the chemical and topological features of small molecules. The similarity is determined by the Tanimoto coefficient (19):
where Na is the number of bits set to 1 in compound a, Nb is the number of bits set to 1 in compound b and Nab is the number of bits common to both, compounds a and b.
Input and output options
There are three ways to start a query with a molecule not included in the database:
Enter SMILES (Simplified Molecular Input Line Entry System)
Draw a molecule using Marvin Sketch
Upload a MOL file using Marvin Sketch
Medical compounds can be retrieved through an expandable ATC-tree, by name, synonym, ATC-code or via known target (name, Uniprot-ID).
The output is a structured table, listing predictions (ATC-codes including confidence interval) containing similarity scores, compound-IDs, molecular structure visualized by Marvin View, target information and physicochemical property intervals of the ATC-group and of the query compound. The score and the color indicate the power of the prediction visualized.
RESULTS
Prediction results
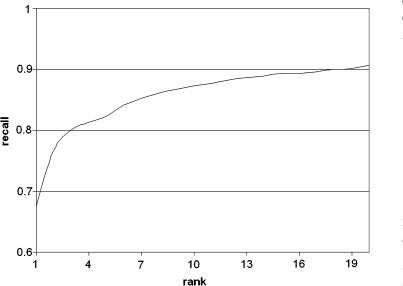
The prediction accuracy is determined by the fraction of correct ATC-class predictions and amounts 67.6%. The distribution of the fractions of correctly predicted indications is shown in Table 1. For a Tanimoto coefficient >0.85 an accuracy of 80.6% is accomplished. A cumulative recall graph is shown in Figure 1. The graph shows the fraction of right predictions of ATC-classes in dependency of the quantity of retrieved structures. By retrieving three molecules the recall gains to about 80% and with 20 retrieved molecules a recall of about 90% is achieved.
Table 1.
Distribution of the fractions of correctly predicted indications
| Range of Tanimoto coefficient | Numbers of hits/misses | Fraction of hits |
|---|---|---|
| 0.4–0.5 | 5/18 | 21.7 |
| 0.5–0.6 | 18/27 | 40.0 |
| 0.6–0.7 | 40/60 | 40.0 |
| 0.7–0.8 | 93/84 | 52.5 |
| 0.8–0.9 | 171/58 | 74.7 |
| 0.9–1.0 | 367/79 | 82.3 |
| 0.0–1.0 | 700/335 | 67.6 |
For the reduced data set of 1035 drugs, 700 right and 335 wrong predictions are investigated. In detail: a similarity score of 90–100% specifies the correct ATC-class in about 82% (367 right and 79 wrong predictions). A hit/miss-rate of about 3/1 is achieved for similarity scores of 70% and higher.
Figure 1.
Cumulative recall for ATC-recognition relative to rank of retrieval.
For the reduced data set of 1035 drugs, the recall is cumulated for one retrieved drug up to twenty retrieved drugs. With three retrieved molecules the recall gains to about 80% and with 20 retrieved molecules a cumulative recall of about 90% is achieved.
Case study
Besides leave-one-out cross-validation statistics, the prediction method was proved by a number of compounds extracted from the SuperTarget database as well as compounds experimentally tested against tumor-cell line assays.
Starting point for the first screening was Enalapril, an ACE-inhibitor, that is used in treatment of hypertension and congestive heart failure. SuperPred identifies six putative drugs having a sufficient similarity to Enalapril indicated by a green color in the result table (Tanimoto coefficient >0.8). An inspection of the referenced literature via the Pubchem-database denoted a similar medical effect for all of them. Table 2 shows exemplarily the names of three of the six putative compounds and the associated reference that describes the medical effect of inhibiting the angiotensin-converting enzyme.
Table 2.
Compounds identified with SuperPred and similar to Enalapril and NSC 600221, respectively
| Name of the compound | Tanimoto coefficient | Medical function | Target protein | Reference |
|---|---|---|---|---|
| Enalapril | 100.00 | ACE-inhibitor | Angiotensin-converting enzyme | (22) |
| Sch 31846a | 94.57 | ACE-inhibitor (predicted) | Angiotensin-converting enzyme (predicted) | (23) |
| Delapril hydrochloride | 83.84 | ACE-inhibitor (predicted) | Angiotensin-converting enzyme (predicted) | (24) |
| Hoe 065b | 81.90 | ACE-inhibitor/increasing central cholinergic activity (predicted) | Angiotensin-converting enzyme (predicted) | (25) |
| NSC 600221c | 100.00 | Antineoplastic agent | Tubulin (predicted) | http://dtp.nci.nih.gov |
| Paclitaxel | 91.62 | Antineoplastic agent | Tubulin beta-1chain | (26) |
a(2S,3aS,7aS)-1-((S)-N-((S)-1-Carboxy-3-phenylpropyl)alanyl) hexahydro-2 indolinecarboxylic acid, 1-ethyl ester, monohydrochloride.
bCyclopenta(c)pyrrole-1-carboxylic acid, 2-(2-((1-(ethoxycarbonyl)-3-phenylpropyl)amino)-1-oxopropyl)octahydro-, octyl ester, (1S-(1-alpha,2-(R*(R*)),3a-beta,6a-alpha))-, (Z)-2-butenedioate (1:1).
The National Cancer Institute Developmental Therapeutics Program (DTP) has screened about 100 000 compounds against a panel of 60 human tumor-cell lines. The results are available on the DTP web site (http://dtp.nci.nih.gov/). The growth inhibition (GI50) and lethal dose (LD50) of the compounds are also retrievable.
Application:
NCI-compound (NSC: 600221) is a screening hit with unverified target. This compound shares a Tanimoto coefficient of 0.92 with the compound Paclitaxel and therefore, it is predicted to be an antineoplastic agent targeting Tubulin.
Enalapril is a well-known ACE-inhibitor. The compound Sch31846 has a similarity of 95% and is supposed to be an ACE-inhibitor, too.
Isolated compounds of the comprehensive tumor-related information resource of the NCI were extracted and screened against the approved drugs included in SuperPred. Many of the screening candidates were characterized by a high physicochemical similarity to well-annotated anti-cancer drugs. For instance, the compound NSC 600221 (Table 2) and the antineoplastic agent Paclitaxel hold a Tanimoto coefficient of 0.91. Both compounds are shown in Figure 2. To analyze the ability to inhibit the proliferation of cancer cells, the GI50-values of both compounds were analyzed with COMPARE, a web accessible tool for investigating mechanisms of cell GI (20). The ability to inhibit the growth of the diverse set of cell lines was highly similar and was indicated by a correlation coefficient of 0.87 calculated by COMPARE. The high correlation coefficient even allowed predictions about the target protein of NSC 600221 (21). As Paclitaxel inhibits microtubule formation by binding to tubulin, the same target came into question for NSC 600221.
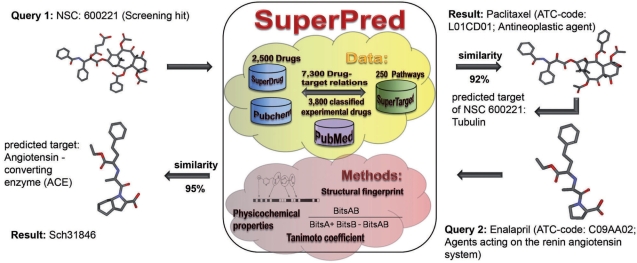
Figure 2.
Assembly of the SuperPred server and possible requests for ATC-code prediction. Data: the SuperPred server now contains 2500 compounds of the SuperDrug database. Additionally, 3800 experimental drugs were classified and stored on the server. The drugs are annotated by 7300 links to targets. Methods: the structural properties of the compounds are stored in so-called structural fingerprints, where each bit encodes for an element of the compound structure. The similarity of two compounds is calculated by using the Tanimoto coefficient. Moreover, physicochemical properties are stored for each compound. SuperPred can be used to find new targets for ligands and vice versa to find new ligands for medical biological targets. There are two possibilities to use the SuperPred server. The figure shows two examples for querying the SuperPred server.
CONCLUSION
The SuperPred web-server was created for predicting medical indications for chemical compounds. The combination of physicochemical property and similarity searching provides the possibility to detect new biologically active compounds and novel targets for drug-like compounds. SuperPred can be applied for drug repositioning purposes, too. A further intention of SuperPred is to find side effects elicited by drugs caused through off-target hits. The use of the web-server is free for all academics.
ACKNOWLEDGEMENTS
The authors wish to thank A. Eckert for assistance during software testing and improvement and S. Struck for critical reading of the article. This work was supported by Deutsche Forschungsgemeinschaft: SFB 449, IRTG Berlin-Boston-Kyoto and Deutsche Krebshilfe. Funding to pay the Open Access publication charges for this article was provided by DFG-Sonderforschungsbereich 449.
Conflict of interest statement. None declared.
REFERENCES
- 1.Voigt JH, Bienfait B, Wang S, Nicklaus MC. Comparison of the NCI open database with seven large chemical structural databases. J. Chem. Inf. Comput. Sci. 2001;41:702–712. doi: 10.1021/ci000150t. [DOI] [PubMed] [Google Scholar]
- 2.Baurin N, Baker R, Richardson C, Chen I, Foloppe N, Potter A, Jordan A, Roughley S, Parratt M, Greaney P, et al. Drug-like annotation and duplicate analysis of a 23-supplier chemical database totalling 2.7 million compounds. J. Chem. Inf. Comput. Sci. 2004;44:643–651. doi: 10.1021/ci034260m. [DOI] [PubMed] [Google Scholar]
- 3.Drews J, Ryser S. The role of innovation in drug development. Nat. Biotechnol. 1997;15:1318–1319. doi: 10.1038/nbt1297-1318. [DOI] [PubMed] [Google Scholar]
- 4.Russ AP, Lampel S. The druggable genome: an update. Drug Discov. Today. 2005;10:1607–1610. doi: 10.1016/S1359-6446(05)03666-4. [DOI] [PubMed] [Google Scholar]
- 5.WHO expert committee. The selection and use of essential medicines. Report of the WHO expert committee, 2005 (including the 14th model list of essential medicines) World Health Organ. Tech. Rep, Ser. 2006;1–119 back cover. [PubMed] [Google Scholar]
- 6.Basak SC, Gute BD, Mills D. Quantitative molecular similarity analysis (QMSA) methods for property estimation: a comparison of property-based, arbitrary, and tailored similarity spaces. SAR QSAR Environ. Res. 2002;13:727–742. doi: 10.1080/1062936021000043463. [DOI] [PubMed] [Google Scholar]
- 7.Liu X, Yang Z, Wang L. Three-dimensional, quantitative-structure-property-relationship study of aqueous solubility for phenylsulfonyl carboxylates using comparative-molecular-field analysis and comparative- molecular-similarity-indices analysis. Water Environ. Res. 2005;77:519–524. doi: 10.2175/106143005x67430. [DOI] [PubMed] [Google Scholar]
- 8.Martin YC, Kofron JL, Traphagen LM. Do structurally similar molecules have similar biological activity? J. Med. Chem. 2002;45:4350–4358. doi: 10.1021/jm020155c. [DOI] [PubMed] [Google Scholar]
- 9.Barbosa F, Horvath D. Molecular similarity and property similarity. Curr. Top. Med. Chem. 2004;4:589–600. doi: 10.2174/1568026043451186. [DOI] [PubMed] [Google Scholar]
- 10.Lagunin A, Stepanchikova A, Filimonov D, Poroikov V. PASS: prediction of activity spectra for biologically active substances. Bioinformatics. 2000;16:747–748. doi: 10.1093/bioinformatics/16.8.747. [DOI] [PubMed] [Google Scholar]
- 11.Poroikov V, Filimonov D, Lagunin A, Gloriozova T, Zakharov A. PASS: identification of probable targets and mechanisms of toxicity. SAR QSAR Environ. Res. 2007;18:101–110. doi: 10.1080/10629360601054032. [DOI] [PubMed] [Google Scholar]
- 12.Geronikaki AA, Lagunin AA, Hadjipavlou-Litina DI, Eleftheriou PT, Filimonov DA, Poroikov VV, Alam I, Saxena AK. Computer-aided discovery of anti-inflammatory thiazolidinones with dual cyclooxygenase/lipoxygenase inhibition. J. Med. Chem. 2008;51:1601–1609. doi: 10.1021/jm701496h. [DOI] [PubMed] [Google Scholar]
- 13.O'Connor KA, Roth BL. Finding new tricks for old drugs: an efficient route for public-sector drug discovery. Nat. Rev. Drug Discov. 2005;4:1005–1014. doi: 10.1038/nrd1900. [DOI] [PubMed] [Google Scholar]
- 14.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004;3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 15.Byvatov E, Fechner U, Sadowski J, Schneider G. Comparison of support vector machine and artificial neural network systems for drug/nondrug classification. J. Chem. Inf. Comput. Sci. 2003;43:1882–1889. doi: 10.1021/ci0341161. [DOI] [PubMed] [Google Scholar]
- 16.Sadowski J, Kubinyi H. A scoring scheme for discriminating between drugs and nondrugs. J. Med. Chem. 1998;41:3325–3329. doi: 10.1021/jm9706776. [DOI] [PubMed] [Google Scholar]
- 17.Gunther S, Kuhn M, Dunkel M, Campillos M, Senger C, Petsalaki E, Ahmed J, Urdiales EG, Gewiess A, Jensen LJ, et al. SuperTarget and Matador: resources for exploring drug-target relationships. Nucleic Acids Res. 2008;36:D919–D922. doi: 10.1093/nar/gkm862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 19.Delaney JS. Assessing the ability of chemical similarity measures to discriminate between active and inactive compounds. Mol. Divers. 1996;1:217–222. doi: 10.1007/BF01715525. [DOI] [PubMed] [Google Scholar]
- 20.Zaharevitz DW, Holbeck SL, Bowerman C, Svetlik PA. COMPARE: a web accessible tool for investigating mechanisms of cell growth inhibition. J. Mol. Graph. Model. 2002;20:297–303. doi: 10.1016/s1093-3263(01)00126-7. [DOI] [PubMed] [Google Scholar]
- 21.Gunther S, Neumann S, Ahmed J, Preissner R. Cellular fingerprints: a novel concept for the integration of experimental data and compound-target-pathway relations. LNBI 4643. 2007:167–170. [Google Scholar]
- 22.Imanishi T, Ikejima H, Tsujioka H, Kuroi A, Kobayashi K, Muragaki Y, Mochizuki S, Goto M, Yoshida K, Akasaka T. Addition of eplerenone to an angiotensin-converting enzyme inhibitor effectively improves nitric oxide bioavailability. Hypertension. 2008;51:734–741. doi: 10.1161/HYPERTENSIONAHA.107.104299. [DOI] [PubMed] [Google Scholar]
- 23.La Rocca PT, Squibb RE, Powell ML, Szot RJ, Black HE, Schwartz E. Acute and subchronic toxicity of a nonsulfhydryl angiotensin-converting enzyme inhibitor. Toxicol. Appl. Pharmacol. 1986;82:104–111. doi: 10.1016/0041-008x(86)90443-6. [DOI] [PubMed] [Google Scholar]
- 24.Fogari R, Malamani G, Zoppi A, Mugellini A, Rinaldi A, Fogari E, Perrone T. Effect on the development of ankle edema of adding delapril to manidipine in patients with mild to moderate essential hypertension: a three-way crossover study. Clin. Ther. 2007;29:413–418. doi: 10.1016/s0149-2918(07)80079-8. [DOI] [PubMed] [Google Scholar]
- 25.Grupp LA, Chow SY. Effects of the novel compound Hoe 065, a central enhancer of cholinergic activity, on voluntary alcohol consumption in rats. Brain Res. Bull. 1991;26:617–619. doi: 10.1016/0361-9230(91)90104-r. [DOI] [PubMed] [Google Scholar]
- 26.Rao S, He L, Chakravarty S, Ojima I, Orr GA, Horwitz SB. Characterization of the Taxol binding site on the microtubule. Identification of Arg(282) in beta-tubulin as the site of photoincorporation of a 7-benzophenone analogue of Taxol. J. Biol. Chem. 1999;274:37990–37994. doi: 10.1074/jbc.274.53.37990. [DOI] [PubMed] [Google Scholar]