Abstract
Rapid alignment of proteins in terms of domains (RAPIDO) is a web server for the 3D alignment of crystal structures of different protein molecules in the presence of conformational change. The structural alignment algorithm identifies groups of equivalent atoms whose interatomic distances are constant (within a defined tolerance) in the two structures being compared and considers these groups of atoms as rigid bodies. In addition to the functionalities provided by existing tools, RAPIDO can identify structurally equivalent regions also when these consist of fragments that are distant in terms of sequence and separated by other movable domains. Furthermore, RAPIDO takes the variation in the reliability of atomic coordinates into account in the comparison of distances between equivalent atoms by employing weighting-functions based on the refined B-values. The regions identified as equivalent by RAPIDO furnish reliable sets of residues for the superposition of the two structures for subsequent detailed analysis. The RAPIDO server, with related documentation, is available at http://webapps.embl-hamburg.de/rapido.
INTRODUCTION
Structural alignment, i.e. the definition of an equivalence map between residues in different structures based on their relative position in space, is a key step in protein structure analysis. The comparison of a protein structure with other structures of the same or similar proteins reveals differences and similarities between related molecules and allows inferring how functional properties are implemented. In the context of a crystallographic structure determination, the alignment of structures of related proteins can identify structurally conserved fragments to be used in molecular replacement (1).
A large number of tools have been developed both for the pairwise and the multiple alignment of structures (2–4). Computer programs for structural alignment can be divided into two main categories depending on whether the molecules under comparison are considered as rigid entities or whether molecular flexibility is taken into account. The first group of computer programs includes DALI (5), CE (6) and MAMMOTH (7) for pairwise alignment and CEMC (8), SSM (9) and MAMMOTH-Mult (10) for multiple alignment. However, it is well known that protein molecules can undergo internal movements, in particular, between their domains and subdomains (11,12). To take molecular flexibility into account, tools for the flexible alignment of protein structures have been implemented. These include FlexProt (13) and FATCAT (14) for pairwise alignment and MultiProt (15) and POSA (16) for multiple alignment.
In this article, we introduce a new web server, named RAPIDO (for rapid alignment of proteins in terms of domains), implementing a new algorithm for the 3D alignment of protein structures in the presence of conformational changes. The web server accepts a set of protein structures and aligns all structures against a reference structure in a pairwise fashion. The algorithm is capable of aligning models of two proteins also in cases of large structural changes such as hinge motions between domains. Furthermore, it is able to identify conformationally invariant regions (rigid bodies) and to produce superpositions.
Among the tools mentioned before, the ones providing the most closely related facilities are FATCAT (http://fatcat.burnham.org/) and FlexProt (http://bioinfo3d.cs.tau.ac.il/FlexProt/). In comparison to these services, RAPIDO has the additional capability of identifying conformational invariant regions when they are not sequential in the residue chain (e.g. when a rigid body contains regions at the N- and C-terminus of a protein separated by another movable rigid body in between). Furthermore, RAPIDO takes into account the variation in the reliability of atomic coordinates by using a B-factor-based weighting scheme. On output, various scripts for displaying the results with PyMOL (http://www.pymol.org) and RasMOL (http://www.umass.edu/microbio/rasmol/index2.htm) are produced.
MATERIALS AND METHODS
Input data
As input, the web server accepts coordinate files in PDB (17) format. The user can either provide the PDB-IDs of structures that are already present in the Protein Data Bank or upload a tarball containing a set of PDB files. The PDB files are parsed and subdivided into chains, which are then called conformers. From the list of conformers, the user can then select a subset for alignment.
Processing method
The structural alignment algorithm consists of four steps:
Detection of short structurally similar fragments, so-called matching fragment pairs (MFPs) (6).
Chaining of the MFPs by a graph-based algorithm.
Identification of rigid bodies with a genetic algorithm (18).
Refinement of the alignment.
At first, the algorithm searches for pairs of structurally similar fragments in the two structures where a fragment is defined as an ungapped stretch of residues and the similarity between fragments is measured by a difference score. The difference score used is the sum over the absolute values of all elements of the difference distance matrix between the Cα-atom positions of the fragments being compared. Pairs of fragments whose difference score is below a defined threshold, are stored as MFPs. In other publications (6,14,19), the term aligned fragment pairs (AFPs) has been used instead of MFPs. In the context of the RAPIDO aligner, we prefer to use the notation of MFPs in order to clarify that in a later stage of the alignment algorithm, a subset of the MFPs forming the initial set is selected to assemble the actual alignment, and the selected MFPs thus become AFPs. In order to do that, the MFPs are represented as nodes of a graph and two MFPs (two nodes) are connected by an edge if they are topologically ordered, i.e. if they are composed of two pairs of fragments that appear in the same order in the two residue sequences. A path in this graph corresponds to a subset of MFPs representing a structural alignment between the two proteins structures. To take into account the varying degree of similarity and size of the MFPs, the gaps between them and their relative displacement, a weight is attached to each edge of the graph in a way inspired by ref. (14). A standard dynamic programing algorithm is then employed to identify the longest path in the graph, which can then be translated into a structural alignment. Further details on the alignment algorithm can be found in (Mosca, Brannetti, Schneider, manuscript in preparation).
Finally, a genetic algorithm originally designed for the identification of conformationally invariant regions in different conformations of the same protein molecule (18) is applied in order to find rigid bodies and the alignment is refined through the application of several heuristics.
Output of the web server
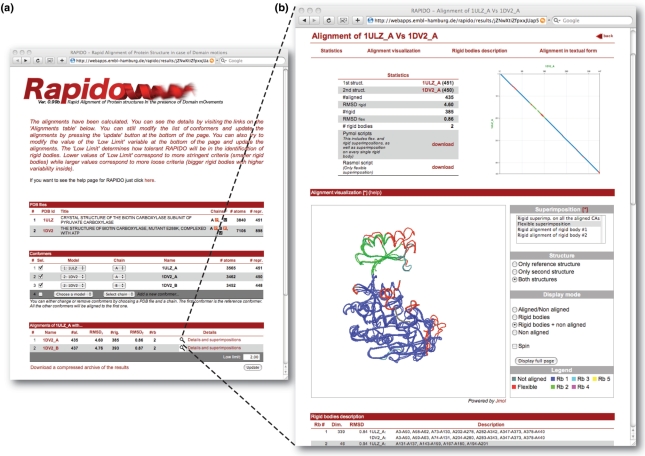
A dot plot of the alignment is provided together with statistics (Figure 1). A textual representation of the alignment is displayed on the web page and can be downloaded in FASTA format. It should be noted that, even if the textual representation of the alignment is referring to the sequence of residues, the equivalent pairs of residues are determined purely on the 3D information contained in two structures. Through a Jmol applet (http://www.jmol.org/), the user can have an immediate overview of the alignment-based superposition. Different types of superpositions are available: rigid superposition on all aligned atoms, superpositions on individual rigid bodies, etc. A particularly revealing way of superposition is the ‘flexible superposition’ of structures. For this type of superposition, the rigid bodies identified in the structural alignment are superimposed separately. For display, parts of the structures falling between the boundaries of two rigid bodies are moved together with the rigid body closest in sequence. The RMSD for a flexible superposition (RMSDf) is calculated as the RMSD over all Cα-atoms of the individual rigid bodies superimposed separately. The superimposed structures in PDB format together with the PyMOL or RasMOL scripts for displaying the superpositions can be downloaded. All output information is color-coded consistently with respect to the rigid body assignments so that conformationally invariant parts can be easily analyzed.
Figure 1.
Output of RAPIDO. (a) The front page displays information about the input data (PDB files and the conformers extracted from them) and a summary of the alignments performed. The selection of conformers or the value of ‘low limit’ can be modified and a new calculation can be submitted by pressing the update button in the lower right corner of the page. (b) Pressing ‘Details and superpositions’ in the table summarizing the alignments will launch a page with more details about the alignment of a particular structure to the reference structure. The page provides various statistics for the alignment, a dot-plot, a Jmol-applet for immediate graphical inspection and a table with a textual description of the results. A consistent color scheme is used for the presentation of all results. Residues assigned to rigid bodies are colored in blue, green, cyan, magenta, etc. (see legend besides the Jmol-window), while residues that were aligned but which were too different to be assigned to a rigid body are colored in red. Residues that were not aligned are colored gray.
AN EXAMPLE: BIOTIN CARBOXYLASE
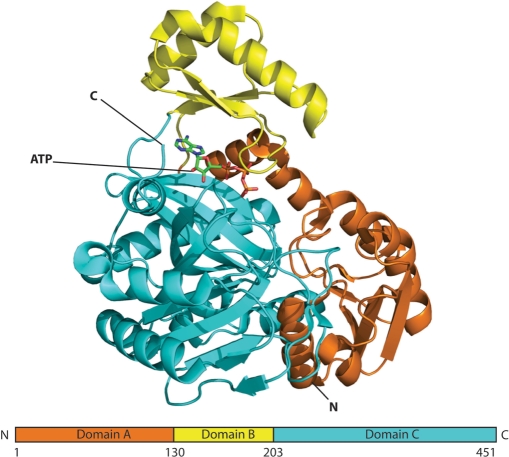
Biotin carboxylase (BC) is a component of enzymes such as pyruvate carboxylase (PC) and acetyl-CoA carboxylase (ACC) mediating the transfer of a carboxyl group through biotin. BCs typically have the ATP-grasp fold (20) and are composed of three sub-domains named A, B and C. The A and C domains form a cylindrical structure and the B domain is positioned at the top of this cylinder creating a pocket in which the active site is located (Figure 2). When ATP binds to the protein, the molecule undergoes a large conformational transition from an open to a closed state in which the B domain moves towards the A and C domains (20,21). Here, we selected two BCs from different organisms, PC from Aquifex aeolicus (22) (PDB-id 1ULZ) and ACC from Escherichia coli (21) (PDB-id 1DV2), in different states (ATP-bound 1DV2 versus apo-1ULZ) to demonstrate the function of RAPIDO.
Figure 2.
Schematic view of BC. Domains A, B and C are color-coded in orange, yellow and cyan, a bound molecule of ATP is shown in stick representation. The figure was created with PyMOL (http://pymol.sourceforge.net).
To start the alignment of the two structures, the PDB-ids of the two crystal structures are filled into the user interface together with an email address to which results will be sent.
After the submission, the web server analyzes the PDB files and subdivides each PDB file into conformers each consisting of one chain. The subset of conformers to be subjected to the alignment procedure is then specified by the user. When the calculations are finished, a link to the URL where the results are stored is sent to the email address provided. This URL contains a randomly generated alphanumeric code to protect the results from unauthorized access. The results remain accessible on the server for 24 hours after the completion of the alignment job. Figure 1 shows the results of the alignment as displayed by the web server. The front page contains a summary table with various statistics: the length (number of residues aligned, #al.), the RMSD of the global superposition (RMSDr), the number of residues belonging to rigid bodies (#rb) and the RMSDf.
Clicking the link on the right side of the rows describing individual alignments in the summary table launches a web page providing more details of the respective alignment. On this page, the first item is a color-coded dot-plot representation (23) of the structural alignment (Figure 1). The 3D superpositions based on the derived alignments can be interactively inspected via a Jmol applet; a set of buttons allows to change the visualization styles, selection of different superpositions modes, the color scheme and the structures actually being displayed.
At the top of the page links are provided for downloading RasMol and PyMOL scripts for the superposition of the structures. Separate PyMOL scripts (pml extension) are generated for each pair of structures and are named with the PBD-id of the two structures followed by a suffix. The suffix indicates the type of superposition: flexible superposition (_flex), rigid superposition (_rigid) and rigid superposition on the i-th rigid body (_rbi). For all the PyMOL scripts rigid bodies and aligned residue can be highlighted by pressing the function keys from F1 to F5 from the PyMOL interface.
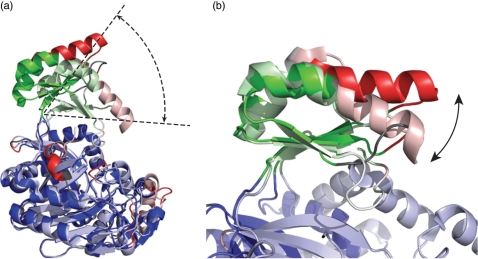
For this example, the first rigid body corresponds to domains A and C and consists of 339 residues that can be superimposed with an RMSD of 0.84 Å. This rigid body is continuous in space but not in polypeptide sequence, containing the N and C terminus but not the central part of the protein sequence. In the center of the polypeptide chain, a short fragment of 46 residues forms a second rigid body, which can be superimposed independently of the rest of the molecule with an RMSD of 0.94 Å and corresponds to a part of the B domain. The flexible superposition (Figure 1) clearly shows that both rigid bodies are structurally very similar although they originate from different conformational states of homologues molecules from different organisms. Superposition of the entire molecules on the Cα-atoms of the first rigid body clearly reveals the displacement between the two conformations of the B domain (Figure 3) depending on the presence or absence of ATP.
Figure 3.
Superpositions of aligned structures of BC from Aquifex aeolicus (1ULZ, light colors) and ACC in E. coli (1DV2, dark colors) (a) Superposition based on 339 Cα-atoms belonging to the first rigid body (in blue). The closure of domain B onto domains A and C is clearly visible. (b) Superposition based on 42 Cα-atoms belonging to the second rigid body (in green). The movement of the helix on top of the B domain (red) with respect to the rest of the B domain (green) becomes visible.
Adjustable parameters
The only user adjustable parameter of the web server is the ‘low limit’—the value for this parameter can be modified in a box displayed at the end of the summary table (Figure 1). This parameter controls to what extent equivalent distances are allowed to change between different models while the corresponding atoms are still counted as belonging to a rigid body (in which in principle all interatomic distances should remain identical). The ‘low limit’ corresponds to the parameter εl used in the comparison of different conformers of the same molecule via a genetic algorithm (18). However, in the present implementation it does not relate to a coordinate uncertainty estimated via Cruickshank's formula (as in ref. 18), but to a more crude weighting function based on B factors only. This choice was made to allow for a fully automatic processing of many PDB-files. The default value for ‘low-limit’ is 2.0 and was optimized for detection of typical domain motions; lower values for ‘low-limit’ will enforce a stricter similarity criterion for distances within rigid bodies leading to smaller rigid bodies, while larger values will do the opposite resulting in fewer rigid bodies of larger size.
CONCLUSION
We have presented a new server for the 3D alignment of protein structures in the presence of conformational changes. The server is able to identify conformational invariant regions between the two structures and to produce superpositions on different rigid bodies separately. Application to a pair of homologues structures of BC from different organisms has shown how the automatic determination of rigid bodies and the distinction between rigid and flexible regions by RAPIDO highlights important functional features of the two analyzed structures. Furthermore, the superposition of the structures on each rigid body separately helps the user identify and quantify the relative movements between conformationally invariant regions.
The choice of the residues for the superimposition is done automatically and based on a sound physical definition of conformationally invariant regions (18) and is not biased by manual intervention.
ACKNOWLEDGEMENTS
This work was supported by grants from Associazione Italiana per la Ricerca sul Cancro (R.M., T.R.S.). Funding to pay the Open Access publication charges for this article was provided by European Molecular Biology Laboratory.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schwarzenbacher R, Godzik A, Jaroszewski L. The JCSG MR pipeline: optimized alignments, multiple models and parallel searches. Acta Crystallogr. 2008;64:133–140. doi: 10.1107/S0907444907050111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lemmen C, Lengauer T. Computational methods for the structural alignment of molecules. J. Comput. Aided. Mol. Des. 2000;14:215–232. doi: 10.1023/a:1008194019144. [DOI] [PubMed] [Google Scholar]
- 3.Sierk ML, Kleywegt GJ. Deja vu all over again: finding and analyzing protein structure similarities. Structure. 2004;12:2103–2111. doi: 10.1016/j.str.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 4.Kolodny R, Koehl P, Levitt M. Comprehensive evaluation of protein structure alignment methods: scoring by geometric measures. J. Mol. Biol. 2005;346:1173–1188. doi: 10.1016/j.jmb.2004.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 6.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 7.Ortiz AR, Strauss CE, Olmea O. MAMMOTH (matching molecular models obtained from theory): an automated method for model comparison. Protein Sci. 2002;11:2606–2621. doi: 10.1110/ps.0215902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guda C, Lu S, Scheeff ED, Bourne PE, Shindyalov IN. CE-MC: a multiple protein structure alignment server. Nucleic Acids Res. 2004;32:W100–W103. doi: 10.1093/nar/gkh464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 10.Lupyan D, Leo-Macias A, Ortiz AR. A new progressive-iterative algorithm for multiple structure alignment. Bioinformatics. 2005;21:3255–3263. doi: 10.1093/bioinformatics/bti527. [DOI] [PubMed] [Google Scholar]
- 11.Gerstein M, Lesk AM, Chothia C. Structural mechanisms for domain movements in proteins. Biochemistry. 1994;33:6739–6749. doi: 10.1021/bi00188a001. [DOI] [PubMed] [Google Scholar]
- 12.Gerstein M, Krebs W. A database of macromolecular motions. Nucleic Acids Res. 1998;26:4280–4290. doi: 10.1093/nar/26.18.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shatsky M, Nussinov R, Wolfson HJ. Flexible protein alignment and hinge detection. Proteins. 2002;48:242–256. doi: 10.1002/prot.10100. [DOI] [PubMed] [Google Scholar]
- 14.Ye Y, Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19(Suppl. 2):II246–II255. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
- 15.Shatsky M, Nussinov R, Wolfson HJ. A method for simultaneous alignment of multiple protein structures. Proteins. 2004;56:143–156. doi: 10.1002/prot.10628. [DOI] [PubMed] [Google Scholar]
- 16.Ye Y, Godzik A. Multiple flexible structure alignment using partial order graphs. Bioinformatics. 2005;21:2362–2369. doi: 10.1093/bioinformatics/bti353. [DOI] [PubMed] [Google Scholar]
- 17.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneider TR. A genetic algorithm for the identification of conformationally invariant regions in protein molecules. Acta Crystallogr. 2002;58:195–208. doi: 10.1107/s0907444901019291. [DOI] [PubMed] [Google Scholar]
- 19.Menke M, Berger B, Cowen L. Matt: local flexibility aids protein multiple structure alignment. PLoS Comput. Biol. 2008;4:e10. doi: 10.1371/journal.pcbi.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tong L, Harwood H.J., Jr. Acetyl-coenzyme A carboxylases: versatile targets for drug discovery. J. Cell Biochem. 2006;99:1476–1488. doi: 10.1002/jcb.21077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thoden JB, Blanchard CZ, Holden HM, Waldrop GL. Movement of the biotin carboxylase B-domain as a result of ATP binding. J. Biol. Chem. 2000;275:16183–16190. doi: 10.1074/jbc.275.21.16183. [DOI] [PubMed] [Google Scholar]
- 22.Kondo S, Nakajima Y, Sugio S, Yong-Biao J, Sueda S, Kondo H. Structure of the biotin carboxylase subunit of pyruvate carboxylase from Aquifex aeolicus at 2.2 A resolution. Acta Crystallogr. 2004;60:486–492. doi: 10.1107/S0907444904000423. [DOI] [PubMed] [Google Scholar]
- 23.Maizel JV, Jr, Lenk RP. Enhanced graphic matrix analysis of nucleic acid and protein sequences. Proc. Natl Acad. Sci. USA. 1981;78:7665–7669. doi: 10.1073/pnas.78.12.7665. [DOI] [PMC free article] [PubMed] [Google Scholar]