Abstract
With over 46 000 proteins, the Protein Data Bank (PDB) is the most important database with structural information of biological macromolecules. PDB files contain sequence and coordinate information. Residues present in the sequence can be absent from the coordinate section, which means their position in space is unknown. Similarity searches are routinely carried out against sequences taken from PDB SEQRES. However, there no distinction is made between residues that have a known or unknown position in the 3D protein structure. We present a FASTA sequence database that is produced by combining the sequence and coordinate information. All residues absent from the PDB coordinate section are masked with lower-case letters, thereby providing a view of these residues in the context of the entire protein sequence, which facilitates inspecting ‘missing’ regions. We also provide a masked version of the CATH domain database. A user-friendly BLAST interface is available for similarity searching. In contrast to standard (stand-alone) BLAST output, which only contains upper-case letters, our output retains the lower-case letters of the masked regions. Thus, our server can be used to perform BLAST searching case-sensitively. Here, we have applied it to the study of missing regions in their sequence context. SEQATOMS is available at http://www.bioinformatics.nl/tools/seqatoms/.
INTRODUCTION
The study into the function and structure of proteins greatly benefits from the availability of tertiary protein structures. Currently, the worldwide Protein Data Bank [wwPDB, (1)] contains over 46 000 entries of proteins and protein/nucleic acids complexes, which are mainly determined by X-ray diffraction. The structure of a large number of proteins is not entirely complete. Often, residues are absent from the determined 3D structure. Thus, these amino acids do not have a determined position in space. The PDB files contain two main records relating to the protein sequence: SEQRES and ATOM. SEQRES contains the protein sequence for which the tertiary structure is reported and the ATOM records contain the atom coordinates. Thus, not all amino acids present in the sequence (SEQRES) necessarily have an entry in the coordinate section of the PDB file.
Protein regions can be absent from the structure for several reasons. It can be not only due to technical problems, but also due to the lack of a fixed tertiary structure. Regions without a fixed tertiary structure, often referred to as disordered regions, can become ordered upon binding to another molecule. Indeed, they have been shown to be important in transcription factors (2) and can be important for protein–protein interaction (3). Besides their biological role, disordered regions are important for crystallographers. Since large disordered regions can complicate crystal formation, constructs for in vitro protein expression are often made such that disordered regions are minimized.
The importance of disordered regions has led to the development of a number of disorder predictors, such as PONDR®, DisEMBL™ (4), GlobPlot™ (5), FoldUnFold (6) and RONN (7) (available at: www.pondr.com, dis.embl.de, globplot.embl.de, skuld.protres.ru/∼mlobanov/ogu/ogu.cgi and www.strubi.ox.ac.uk/RONN). In addition, a database of protein disorder [DisProt, (8)] has been developed that provides curated information on disorder.
However, not all ‘missing’ residues are disordered. Additional (experimental) evidence is needed to conclude that a ‘missing’ region is disordered. Combining disorder predictions with information on missing regions from previously crystallized homologous proteins may be beneficial to the development of protein expression constructs for crystallography. We have developed SEQATOMS in order to provide an overview of all missing regions. Indeed, we do not focus on disordered regions, but on missing regions. SEQATOMS consists of number of sequence databases and several services, including BLAST and keyword searches, to access the sequence information. The first database is based on PDB and the second database is based on the CATH domain database (9). Although it is derived from PDB, we included CATH since it provides protein domains and structure classification. All residues missing in the (PDB) coordinate section are indicated by lower-case letters. For completeness, PDB SEQRES and DisProt are also included. To facilitate the sequence analysis of missing regions in their sequence context and, specifically, to illustrate the use of case-sensitive BLAST alignments, we provide a BLAST (10) web server that makes it possible to retain lower-case letters in the BLAST results. Thus, the user can see at a glance, which regions of the provided query protein correspond with regions without determined structure in PDB. Here, we show its application to the sequence analysis of missing amino acid regions in protein structures.
METHODS
Database construction
The web server currently provides four protein sequence databases: PDB SEQATOMS, CATH, PDB SEQRES and DisProt. The first two are constructed as described below, the third contains all sequences from the PDB SEQRES records and is downloaded from PDB (pdb_seqres.txt). For the fourth, the DisProt (8) FASTA file (www.disprot.org) was processed to lower-case-mask the disordered regions and the protein name, synonyms and organism name were added to the FASTA header.
Seqatoms
SEQATOMS is derived from PDB (1). For this database, we process all protein and protein/nucleic acids complexes in PDB. The PDB macromolecular Crystallographic Information File (mmCIF) format already contains an alignment of the amino acid residues from SEQRES and ATOM records under the ‘_pdbx_poly_seq_scheme’ item. Residues absent from the coordinate section are marked with ‘?’ in this scheme. The residues are converted to a FASTA sequence and all positions marked with ‘?’ are converted to lower-case letters. Thus, sequences without missing residues remain present. Next, all sequences shorter than three residues or matching only ‘X’ are filtered out. For the remaining sequences a FASTA definition line is produced as follows. The FASTA ID is the name of the PDB entry (taken from _entry.id) to which the chain identifier is appended. The FASTA description starts with an upper-cased part, which contains the text from ‘_struct_keywords.pdbx_keywords’ item. In addition, the text for the structure description, provided by ‘_struct.title’ or ‘_struct.pdbx_descriptor’, is added. After completion, the database is made non-redundant case-sensitively. Hence, proteins with identical sequences (ignoring case), which miss different regions in their determined structure, are not regarded as identical. FASTA headers of redundant sequences are concatenated to be able to view the descriptions of the ‘cluster’ members.
Cath
Since its previous release (v3.0.0; August 2006), CATH (9) provides the domain sequences as ATOM, COMBS FASTA files. The COMBS sequences provide the full sequence instead of only the residues present in the ATOM records. We use the Levenshtein algorithm to align the FASTA sequences in the CATH ATOM file with the corresponding one in the COMBS file (www.cathdb.info; version 3.1.0). The gaps in the ATOM-based sequence are replaced with the corresponding lower-cased residues in the COMBS sequence.
As before, the FASTA database is made non-redundant case-sensitively and CATH IDs are concatenated. As the FASTA sequences do not contain any description, we produce a FASTA description line containing the CATH class, architecture, topology and homology data present in the domain description file (CathDomainDescriptionFile.v3.1.0).
Web server implementation
Input
The user must provide a FASTA protein sequence and may select the following parameters: BLAST database (masked PDB SEQATOMS, masked CATH, masked DisProt or the original PDB SEQRES database), masking character, masking colour and a number of BLAST parameters.
Case-sensitive BLAST implementation
The case-sensitive alignment output is produced by post-processing the (plain-text) BLAST report with Perl and BioPerl (Bio::SearchIO) (11). Post-processing is required, since formatdb, which produces the BLAST databases, converts all letters to upper case. The start, stop and ID of all BLAST hits are taken from the report and the corresponding sequence strings are extracted from our databases. The similarity line is lower-case masked and the new similarity and hit strings replace the original strings. BioPerl's Bio::Graphics::Panel module is used for the generation of the alignment graphic. BLAST output in XML format is parsed with regular expressions. The hit strings and similarity strings are replaced with their masked versions.
Output
The result page is a reformatted BLAST output in pair-wise or XML format. Regions absent from the determined structure are indicated by lower-case letters or by ‘x’ and coloured depending on user input. The pair-wise output contains a graphic showing the hit distribution on the query sequence and provides links to extract the complete sequences from our databases. Moreover, all hits are linked to their source databases, NCBI Entrez Protein, CATH and PDB. The XML output, provided to support automated access, is a normal BLAST XML output, in which hit and similarity strings have been replaced with the corresponding masked strings.
RESULTS AND DISCUSSION
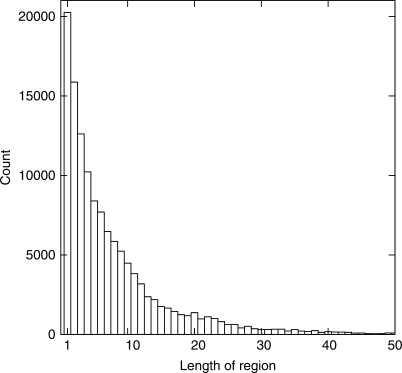
The wwPDB (1) stores 3D molecular structures in three formats (1,12,13): PDB, mmCIF, PDBML/XML. The mmCIF format contains an alignment of the residues in the sequence and in the coordinate sections. We have processed these mmCIF files to produce the PDB SEQATOMS database. All residues that are absent from the structure are indicated by lower-case letters. The resulting sequence database has been made non-redundant case-sensitively to be able to show the variation of ‘missing’ regions on otherwise identical sequences. This variation is indeed quite large as this database is 59% larger when it is made non-redundant in a case-sensitive way as compared to a case-insensitive way. The PDB contained 109 205 proteins or protein chains (longer than three residues, containing at least one amino acid character other than ‘X’; numbers from 16 January 2008). As many as 64% of these proteins had at least one missing, or lower-case masked, residue and 26% had at least ten missing residues. Figure 1 shows the distribution of all missing regions of a certain length in our PDB-derived database (redundant version). Please, note that one chain can have more than one missing region. Mainly, regions at the begin or the end of a protein sequence are missing from the structures.
Figure 1.
Histogram of missing residues in PDB protein structures. All missing regions are counted. This count is larger than the number of chains, since a single chain can have several missing regions.
For completeness, we added the sequences from DisProt, PDB SEQRES and the CATH domain database to our web server. All 93 885 domains in CATH version 3.1.0 have been processed. After the introduction of residues absent from the ATOM-based sequence into this ATOM-based sequence, the database was made non-redundant case-sensitively. This resulted in a CATH-derived database that is 33% larger as compared to the same database made non-redundant case-insensitively. The addition of CATH (class, architecture, topology and homology) information to the FASTA sequence facilitates interpreting the BLAST results.
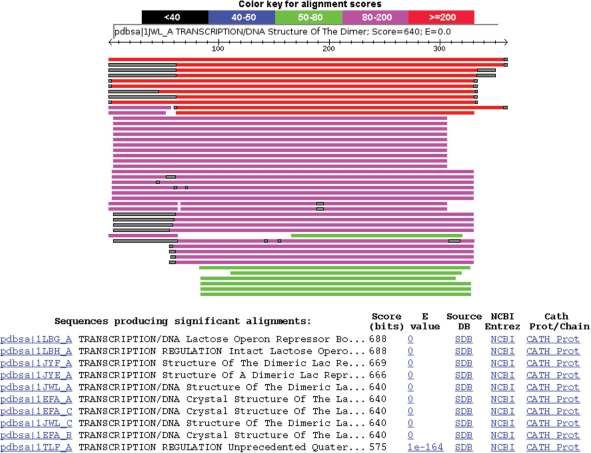
The web interface provides the possibility to carry out BLAST searches against the databases. The user can select the masking character (lower-case or ‘x’) and masking colour (black, grey, red), similar to the NCBI web-server. However, the masking at NCBI refers to low-complexity regions, not to lower-case regions present in the database. The pair-wise (HTML) output provides an alignment graphic to visualize the overall missing regions. This graphic shows the distribution of the hit sequences over the query as well as the lower-case (missing) regions that align with query residues (Figure 2). All missing regions, including those that align with query gaps, are indicated in the alignments (Figure 3). To facilitate the retrieval of additional information, the pair-wise BLAST output provides links to the sequences in our databases, as well as links to the source databases, Entrez Protein, CATH and PDB. In addition to BLAST searches, the user can search proteins by keyword and IDs. For users interested in disorder, we provide links to several disorder predictors. Automated access to the sequence entries as well as to keyword and BLAST searches is provided via URLs or scripts as described on the ‘Services’ web page. Especially for automated access, the XML output option is provided. The important difference with standard BLAST XML output is the presence of masked regions in the hit and similarity strings.
Figure 2.
An example BLAST output for the query 1LBG_A (Lactose Operon Repressor). The lower-case regions are indicated in the alignment graphic (grey) and in the alignment (Figure 3). The description section of the BLAST output provides links to the selected sequence database(s), the source database, NCBI Entrez Protein and CATH.
Figure 3.
An example alignment showing lower-case masking (red) of the second hit sequence (1LBH_A) from the BLAST output presented in Figure 2. The FASTA header has been truncated and only the first line of the alignment is shown here.
To exemplify a possible use of this server, we search for missing regions in 1LBG_A, a lactose operon repressor. This protein has three missing residues at the C-terminus (358–360). However, the BLAST result, using PDB SEQATOMS and DisProt, shows several highly significant hits with more missing residues. A DisProt hit (DP00433) has disordered regions from position 1–62 (six regions in total) and a PDB SEQATOMS hit, 1JYF_A, has missing residues from 1–61 and 334–349. Disorder prediction using RONN (see ‘Disorder’ web page) indicates residues 22–38 and 325–360 are disordered. This server thus provides an overview of overall missing regions (Figure 2) in similar or homologous sequences and the links provide a convenient way to retrieve additional information to evaluate these missing regions.
CONCLUSION
The presented web server visualizes BLAST results case sensitively and the alignment graphic provides an overview of missing regions. As the server post-processes the BLAST results, the implementation is independent of BLAST releases. In addition, it is relatively straightforward to produce pair-wise (HTML) output for a variety of similarity search programs already available in BioPerl's Bio::SearchIO, such as WU-BLAST and FASTA. Mainly for automated access, a BLAST XML output is provided that shows the lower-cased regions in the similarity and hit strings. Automated BLAST or keyword searching and sequence retrieval are supported.
ACKNOWLEDGEMENTS
This work was supported by ENFIN, a Network of Excellence funded by the European Commission within its FP6 Programme, under the thematic area ‘Life sciences, genomics and biotechnology for health’, contract number LSHG-CT-2005-518254. We would like to thank Drs Rachel Kramer Green and Zukang Feng (from RCSB PDB) for providing feedback with respect to PDB mmCIF issues and Dr Gary Benson for his suggestion to improve the alignment graphic. Open Access charges for this article were waived by NAR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 4.Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galzitskaya OV, Garbuzynskiy SO, Lobanov MY. FoldUnfold: web server for the prediction of disordered regions in protein chain. Bioinformatics. 2006;22:2948–2949. doi: 10.1093/bioinformatics/btl504. [DOI] [PubMed] [Google Scholar]
- 7.Yang ZR, Thomson R, McNeil P, Esnouf RM. RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics. 2005;21:3369–3376. doi: 10.1093/bioinformatics/bti534. [DOI] [PubMed] [Google Scholar]
- 8.Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, Szabo B, Tompa P, Chen J, Uversky VN, et al. DisProt: the Database of Disordered Proteins. Nucleic Acids Res. 2007;35:D786–D793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greene LH, Lewis TE, Addou S, Cuff A, Dallman T, Dibley M, Redfern O, Pearl F, Nambudiry R, Reid A, et al. The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res. 2007;35:D291–D297. doi: 10.1093/nar/gkl959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JGR, Korf I, Lapp H, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westbrook J, Ito N, Nakamura H, Henrick K, Berman HM. PDBML: the representation of archival macromolecular structure data in XML. Bioinformatics. 2005;21:988–992. doi: 10.1093/bioinformatics/bti082. [DOI] [PubMed] [Google Scholar]
- 13.Westbrook J, Feng Z, Jain S, Bhat TN, Thanki N, Ravichandran V, Gilliland GL, Bluhm W, Weissig H, Greer DS, et al. The Protein Data Bank: unifying the archive. Nucleic Acids Res. 2002;30:245–248. doi: 10.1093/nar/30.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]