Abstract
The major public microarray repositories Gene Expression Omnibus and ArrayExpress are growing rapidly. This enables meta-analysis studies, in which expression data from multiple individual studies are combined. To facilitate these types of studies, we developed Microarray Retriever for searching and retrieval of data from GEO and ArrayExpress. The tool allows access to the two repositories simultaneously, to search in the repositories using complex queries, to retrieve microarray data for published articles and to download data in one structured archive. The tool is available on the web at: http://www.lgtc.nl/MaRe/
INTRODUCTION
Microarray technology is now routinely used for genome-wide mRNA expression and epigenetic profiling. As a consequence, there is a quickly growing amount of microarray data made available in public databases. This may improve the interpretation of new experimental studies through comparison with data already publicly available, and allow for large-scale meta-analysis. Use of data in the repositories gives increased statistical power, and facilitates confirmation of hypotheses coming from one study with data from another study, identification of artifacts and separation of primary effects from secondary (1). For example, meta-analysis of microarray data was useful for identification of common transcriptional profiles of neoplastic transformation and progression (2), discovery of genes disproportionately overexpressed in specific cancer types (3), construction of robust high-resolution gene coexpression networks (4), and identification of rhythmically expressed genes in Drosophila (5).
There are many hurdles to be taken when integrating data from different studies, such as the necessity to map the probes on different arrays to transcripts, the necessity to account for different experimental designs and analysis algorithms, and to retrieve the relevant datasets from microarray data repositories. The latter is addressed by MaRe, the tool that we developed.
Currently, there are no public software tools, which provide combined and interactive access to the main microarray data repositories. Microarray Retriever (MaRe) facilitates meta-analysis by enabling searching and batch data retrieval from the two major public microarray repositories: Gene Expression Omnibus (GEO, National Center for Biotechnology Information; 6,7) and ArrayExpress (AE, European Bioinformatics Institute; 8,9). MaRe allows the user to search these repositories for experiments with accession numbers, authors, species, date of submission, array platform and keyword search terms. In addition, users can first search PubMed on authors and keywords for relevant literature and then search GEO and ArrayExpress for experimental data associated with this literature. Alternatively, a custom list of PubMed IDs can be uploaded to obtain data for specific articles known in advance. After the search is complete, MaRe enables downloading of selected results in one step. This avoids the time-consuming procedure of manual and sequential downloading from the web or ftp sites of the repositories. All retrieved results are stored using a clear directory structure.
METHODS
Implementation
MaRe is written in Perl. Searches in GEO are performed remotely by sending queries to NCBI E-Utilities (10). A 3-s delay between E-Utilities calls is implemented according to the NCBI guidelines. Search in ArrayExpress is done locally by parsing the ArrayExpress annotation XML file. The file is downloaded from the EBI during a search if the previous version of the file is older than 1 h (http://www.ebi.ac.uk/arrayexpress/q-aer/).
Search options
The MaRe web interface contains three boxes for input of the query terms: ‘Accessions (A)’, ‘Authors/keywords (B)’ and ‘Species/date/platform (C)’.
Box A accepts accession numbers of GEO experiments (Series and Datasets), accession numbers of ArrayExpress experiments and PubMed IDs of papers potentially associated with experimental data in either of the repositories.
Box B specifies authors and keywords to be searched for in the meta-data present in the microarray repositories and/or PubMed. The queries can contain logical operators, brackets and quotes (a detailed description is provided in the online help file). Approximately two-thirds of entries in GEO and AE are linked to PubMed. Therefore, PubMed searches should be preferentially used in combination with searches in annotation.
Box C enables searching on or limiting searches on specific species (for example, ‘Mus musculus’ AND ‘Rattus norvegicus’—to search for experiments where both species were examined), date of submission to the repository (can be an interval) and platforms (using platform accession numbers or platform keywords).
In the keywords field (box B) and the platform keywords field (box C), Entrez limits can be added to the input terms, e.g. ‘p53[Title]’. The limits are only used for searches in GEO and PubMed.
Three additional boxes are provided for configuring the search. Box ‘Query logics’ is used to define how the boxes A, B and C should be combined to generate the complete query. Box ‘Search options’ contains the following options:
to search for experiments and the associated platforms or to search for platforms only;
to search in GEO and/or in ArrayExpress;
to retrieve Series and/or Datasets from GEO;
to retrieve all experiments from ArrayExpress or only those, which do not duplicate data already retrieved in the GEO search;
to retrieve raw data for experiments or to retrieve only the processed data.
An email address should be entered in the ‘Start search’ box before the search can be started. MaRe will send a notification with the URL of the data archive to this email address.
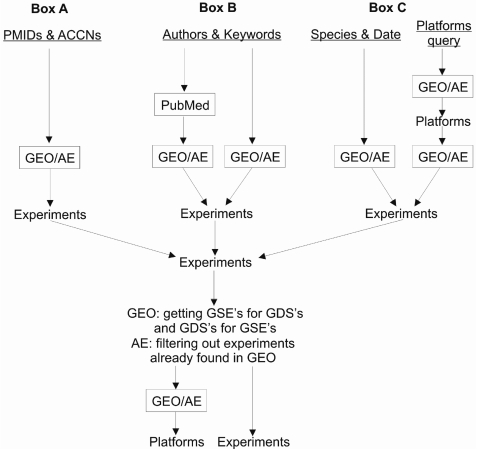
Searches based on box A, B and C inputs are performed independently. Subsequently, the results are combined according to the user-defined logics and redundant hits are removed. The search structure is further outlined in Figure 1.
Figure 1.
Overview of the search scheme applied. Search on each of the underlined input terms is performed independently, after which the results are combined. After obtaining Series on Datasets and Datasets on Series for GEO and filtering out experiments already found in GEO for AE, platforms are retrieved for the identified experiments. The experiments and the platforms are shown on the results page.
Extra features
When searching with GEO accession numbers, the user can select ‘Retrieve GSE and GDS’. Doing so, series associated with the found datasets and datasets associated with the found series are automatically retrieved. This is similar to the search logics implemented in GEO Entrez. If ‘Retrieve only GSE’ or ‘Retrieve only GDS’ is chosen, entries which meet the query themselves are retrieved (series or datasets, respectively).
When searching ArrayExpress, the user can select to retrieve only data ‘Not retrieved from GEO’. In that case, AE experiments that duplicate GEO entries already found in the search will not be displayed on the results page. Primary accession numbers and secondary accession numbers of AE experiments are used to distinguish those that are also found in GEO. Primary accession numbers of such experiments have the specific format ‘E-GEOD-number’, where the number indicates the corresponding GSE, secondary accession numbers coincide with the GSE accession numbers in GEO.
When platform terms are entered in an ‘Experiments and platforms’ search, the platforms are found first. Subsequently, experiments associated with them are retrieved and combined with experiments found for boxes A and B. Platforms in the results page are those which correspond to the found experiments. Advantages of using platform terms in a search for ‘Experiments and platforms’ are that the experimental data retrieved will be limited to a specific platform, and that only those platforms will be displayed on which experiments have been performed (function not available in GEO). When a search for ‘Platforms only’ is performed, the repositories are searched for platforms, which meet the platform terms and all platforms, including those with no associated experiments, are displayed in the results page.
Downloading
As soon as some of the search results are selected for downloading and the downloading job is started, an email message is sent to inform the user that the job has been started. Processed data from GEO are downloaded in GEO SOFT format (6). Processed data from the ArrayExpress ftp site are downloaded in all available formats (8). If the user has chosen to download raw data, these data are downloaded as well.
MaRe keeps the load on the GEO and ArrayExpress servers minimal by downloading all data on a per file basis instead of downloading all files at once. While downloading, files are retrieved from multiple directories on the ftp servers of the repositories and organized locally so that files for the same experiment are placed in the same folder. All archives are unzipped. After the downloading job is complete, the downloaded files are zipped into a single archive. Finally, an email is sent to the user with the URL to the archive on the MaRe server.
RESULTS AND DISCUSSION
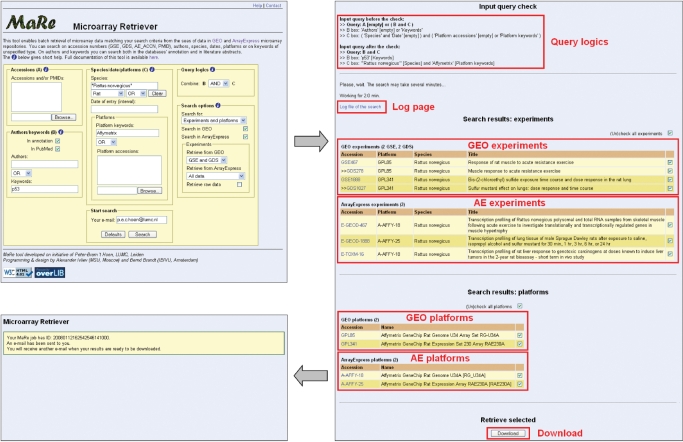
An example of a search is provided in Figure 2. The search parameters are configured within the user-friendly interface. The layout of the results page is compact and gives a good overview with hyperlinks to the entries in GEO or AE.
Figure 2.
Example of a search. Search parameters are configured on the first page, entries to download are chosen on the next page after the search is complete, ID of the accepted downloading job is finally reported. The red boxes and text indicate the different sections of the results page.
To illustrate the performance of our application, we retrieved all data from GEO and ArrayExpress connected with a particular area of research—ovarian cancer. A query on keyword ‘ovarian[Title] and (cancer[Title] or tumour*[Title] or tumor*[Title])’ (box B) limited to the species Homo sapiens (box C) in the annotations of the two repositories and in PubMed resulted in 16 GEO Series (29 platforms) and 13 ArrayExpress experiments (19 platforms). The search took 2 min. Processed data for all experiments and complete descriptions of all associated platforms were selected for download. Downloading all data to the MaRe server took 2 h and 50 min. Retrieving the complete archive from the MaRe server using the emailed URL took 2 h and 30 min. The size of the archive was 2.5 Gb, while the size of the unzipped data comprised 5.5 Gb.
As stated earlier, keyword searches in GEO and ArrayExpress can be performed in the meta-data of the repositories and/or PubMed. Search in PubMed can result in relevant entries not found by an equivalent search in the GEO annotation fields. For example, searching for GEO Series with the keyword query ‘p53[Title] or tp53[Title]’ yielded 30 GEO Series (as on 12 January 2008). Searching for GEO Series with the same query via PubMed added 11 more experiments. Four of these 11 experiments (GSE2155, GSE3072, GSE7678 and GSE8023) contain neither of the terms ‘p53’ or ‘tp53’ in their titles nor in the GEO annotation (except for the ‘citation’ field which is not queried via GEO Entrez). Nevertheless, all of these experiments were manually checked to be relevant for p53 research. In this way, searching in PubMed increases the recall of the search.
Microarray Retriever provides the following options, which are not available in GEO and ArrayExpress:
simultaneous access to both repositories;
search in the repositories via PubMed;
selection of platforms on which experiments have actually been done;
batch downloading of entries.
Besides MaRe, there are several other tools which have an option of downloading microarray data from public sources: GEOquery (11), SeqExpress (12) and ArrayQuest (13). All of them are limited to GEO, MaRe being the only tool which combines both of the major public microarray repositories. MaRe is also the only tool which offers robust searching functions using diverse types of parameters and keywords. Another MaRe-specific function is search in the microarray repositories via PubMed. Finally, it is currently the only existing tool specifically designed for searching and downloading of public microarray data to a local machine. MaRe is therefore a valuable addition to the existing tools, which are primarily designed for data analysis.
CONCLUSION
In the coming years, large-scale meta-analysis of microarray data will become more and more important. Our tool facilitates this type of analysis by querying and automatic downloading of relevant data from the most frequently used microarray data sources in the public domain.
ACKNOWLEDGEMENTS
This study was initiated and partially funded by the LUMC-Moscow State University Cooperative Program on Bioinformatics. P.A.C.'tH. was supported by a VENI-grant from the Dutch Organization for Scientific Research (NWO grant 2005/03808/ALW). B.B. was supported by ENFIN, a Network of Excellence funded by the European Commission within its FP6 Programme, under the thematic area ‘Life sciences, genomics and biotechnology for health’, contract number LSHG-CT-2005-518254. The Open Access charges for this paper were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Larsson O, Wennmalm K, Sandberg R. Comparative microarray analysis. OMICS. 2006;10:381–397. doi: 10.1089/omi.2006.10.381. [DOI] [PubMed] [Google Scholar]
- 2.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc. Natl Acad. Sci. USA. 2004;101:9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Barrette TR, Ghosh D, Chinnaiyan AM. Mining for regulatory programs in the cancer transcriptome. Nat. Genet. 2005;37:579–583. doi: 10.1038/ng1578. [DOI] [PubMed] [Google Scholar]
- 4.Jupiter DC, Vanburen V. A visual data mining tool that facilitates reconstruction of transcription regulatory networks. PLoS ONE. 2008;3:e1717. doi: 10.1371/journal.pone.0001717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keegan KP, Pradhan S, Wang JP, Allada R. Meta-analysis of Drosophila circadian microarray studies identifies a novel set of rhythmically expressed genes. PLoS Comput. Biol. 2007;3:e208. doi: 10.1371/journal.pcbi.0030208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R. NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edgar R, Domrachev M, Lash AE. Gene expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parkinson H, Kapushesky M, Shojatalab M, Abeygunawardena N, Coulson R, Farne A, Holloway E, Kolesnykov N, Lilja P, Lukk M, et al. ArrayExpress—a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007;35:D747–D750. doi: 10.1093/nar/gkl995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brazma A, Parkinson H, Sarkans U, Shojatalab M, Vilo J, Abeygunawardena N, Holloway E, Kapushesky M, Kemmeren P, Lara GG, et al. ArrayExpress—a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2003;31:68–71. doi: 10.1093/nar/gkg091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sean D, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 12.Boyle J. Gene-expression Omnibus integration and clustering tools in SeqExpress. Bioinformatics. 2005;21:2550–2551. doi: 10.1093/bioinformatics/bti355. [DOI] [PubMed] [Google Scholar]
- 13.Argraves GL, Jani S, Barth JL, Argraves WS. ArrayQuest: a web resource for the analysis of DNA microarray data. BMC Bioinform. 2005;6:287. doi: 10.1186/1471-2105-6-287. [DOI] [PMC free article] [PubMed] [Google Scholar]