Abstract
Structural details of protein–protein interactions are invaluable for understanding and deciphering biological mechanisms. Computational docking methods aim to predict the structure of a protein–protein complex given the structures of its single components. Protein flexibility and the absence of robust scoring functions pose a great challenge in the docking field. Due to these difficulties most of the docking methods involve a two-tier approach: coarse global search for feasible orientations that treats proteins as rigid bodies, followed by an accurate refinement stage that aims to introduce flexibility into the process. The FireDock web server, presented here, is the first web server for flexible refinement and scoring of protein–protein docking solutions. It includes optimization of side-chain conformations and rigid-body orientation and allows a high-throughput refinement. The server provides a user-friendly interface and a 3D visualization of the results. A docking protocol consisting of a global search by PatchDock and a refinement by FireDock was extensively tested. The protocol was successful in refining and scoring docking solution candidates for cases taken from docking benchmarks. We provide an option for using this protocol by automatic redirection of PatchDock candidate solutions to the FireDock web server for refinement. The FireDock web server is available at http://bioinfo3d.cs.tau.ac.il/FireDock/.
INTRODUCTION
Protein–protein interactions are essential for many biological processes in the cell. In recent years, there has been a major progress in high-throughput identifications of interacting proteins (1). Despite recent advances in the Structural Genomics project (2), the number of experimentally solved structures of protein–protein complexes remains very low (3). Therefore, computational methods are essential for 3D characterization of protein–protein interactions.
Protein–protein docking aims to predict the 3D structure of a protein complex given the structures of the individual proteins that assemble it. Protein flexibility (4) and the absence of robust scoring functions present a major challenge in the docking field. The flexibility of proteins includes both backbone and side-chain movements, and dealing with it significantly extends the search space for the optimal complex structure (5,6). In addition, state of the art scoring functions are not tolerant enough to the inaccuracies in the predicted structures (7). Therefore, near-native solutions may not be highly ranked. Due to these difficulties, many docking methods apply a two-stage approach. First, soft rigid docking is performed (8–11), allowing a certain amount of steric clashes. Then, thousands of solution candidates are refined and re-scored in a refinement stage (5,6). In this stage, each solution candidate is improved by modeling the protein flexibility and optimizing the rigid-body orientation.
Most of the refinement methods use an optimization of the side-chain conformations and/or rigid-body orientation in order to minimize a certain scoring function. Only a few of them incorporate backbone flexibility. One of the main differences between the refinement methods is in the approach used for side-chain optimization: Monte-Carlo (MC) (12,13), graph theory-based methods (14,15), linear programming (LP) (16), etc. The optimization of the rigid-body orientation usually involves a randomized perturbation followed by energy minimization. The minimization is performed using methods such as Steepest Descent (17), Newton–Raphson (18,19), Quasi-Newton (12,16) and more.
There are several web servers that deal with different aspects of the docking challenge: ZDock (20) and PatchDock (21) perform rigid-body docking. ClusPro (22,23) can filter, cluster and rank docking solution candidates. SmoothDock (24) is a newer version of ClusPro that also refines the representatives of the largest clusters. RosettaDock (12,25) web server allows local search in the vicinity of a single given input solution candidate. The GRAMM-X (26) and Hex (27) web servers perform rigid-body docking followed by an optimization of the rigid-body orientation.
FireDock method for flexible refinement and scoring of protein–protein docking solutions was developed by Andrusier et al. (16) and was previously available only for downloading. Here we present the FireDock web server that provides a user-friendly interface for running this protocol online. It is the first web server for refinement of protein–protein docking solutions that includes a side-chain optimization component. It allows a high-throughput refinement of up to 1000 solution candidates. The method simultaneously targets the problem of flexibility and scoring of solutions produced by fast rigid-body docking algorithms. The output provides a list of refined complexes, sorted by a binding energy function, and a 3D visualization for observing and comparing between the refined complexes.
Additionally, we suggest a platform for performing full docking protocol: rigid-body docking by the PatchDock web server (21) and a redirection of the results for refinement and scoring by FireDock web server. This protocol was thoroughly tested (16) on protein–protein docking benchmarks (28,29).
THE FIREDOCK METHOD
The input to the FireDock algorithm is a set of candidate complexes. Each complex consists of two proteins: receptor and ligand. The method refines each candidate and ranks all the candidates according to the binding energy.
The FireDock (16) method includes three main steps:
Side-chain optimization: The side-chain flexibility of the receptor and the ligand is modeled by a rotamer library. The optimal combination of rotamers for the interface residues is found by solving an integer LP problem (30–32). This LP minimizes a partial energy function consisting repulsive van der Waals and rotamer probabilities terms.
Rigid-body minimization: This minimization stage is performed by a MC technique that attempts to optimize an approximate binding energy by refining the orientation of the ligand structure. The binding energy consists of softened repulsive and attractive van der Waals terms. In each cycle of the MC method, a local minimization is performed by the quasi-Newton algorithm (33,34). By default, 50 MC cycles are performed.
Scoring and ranking: This final ranking stage attempts to identify the near-native refined solutions. The ranking is performed according to a binding energy function that includes a variety of energy terms: desolvation energy (atomic contact energy, ACE), van der Waals interactions, partial electrostatics, hydrogen and disulfide bonds, π-stacking and aliphatic interactions, rotamer's; probabilities and more.
The FireDock method was extensively tested (16) on docking candidates generated by the PatchDock method (10,21) for cases from benchmark 1.0 and 2.0 (28,29). FireDock succeeded in ranking a near-native solution in the top 15 predictions for 83% of the 30 enzyme–inhibitor test cases and for 78% of the 18 semi-unbound antibody–antigen test cases.
FIREDOCK WEB SERVER
The FireDock web server is available at http://bioinfo3d.cs.tau.ac.il/FireDock/.
Input
There are two input options for the FireDock web server. In the first option, the user uploads or specifies codes of two Protein Data Bank (PDB) (35) files (receptor and ligand) and provides a list of up to 1000 transformations (the required format is detailed in the Help page of the web server). Each transformation, when applied on the ligand, produces a candidate docking solution. In the second option, the user can upload an input PDB file, with each docking solution represented by a MODEL. The candidate solutions for FireDock can be generated by rigid-body docking methods, such as PatchDock (10,21), methods that use Fast Fourier Transform (FFT) such as ZDOCK (20,36), GRAMM-X (26), Hex (27), etc. The PatchDock server (http://bioinfo3d.cs.tau.ac.il/PatchDock/) includes an option for automatic redirection of the solution candidates to FireDock.
In addition, the user may modify the ‘Number of output structures’ parameter. This parameter determines the number of best scoring candidates for which a PDB file with the refined structure will be generated. The server allows generating up to 100 PDB files. A link to the output web-page is sent to the email of the user as soon as the refinement process is finished.
The server includes optional advanced parameters for adjusting the refinement and scoring for a specific biological system. The user can specify the type of the complex (Default, Antibody–Antigen or Enzyme–Inhibitor), which is used for adjusting the weights of the scoring function. Furthermore, the user can specify if the proteins are in their bound or unbound conformation and if certain side-chains are known to be flexible or fixed.
Other advanced parameters determine the level of refinement. The ‘Restricted’ refinement mode allows only the clashing residues to be flexible. The ‘Full’ refinement mode allows all the interface residues to be flexible and uses an extended rotamer library. We recommend using the restricted mode at first, for coarse refinement, and to use the full mode on the final best candidates. The user can also set the number of rigid-body optimization cycles. This parameter influences the range of rigid-body movements around each original solution candidate. Finally, the user can scale the atomic radii used in energy calculations. This parameter influences the extent of acceptable steric clashes in the final refined solutions. We recommend using 0.8 for coarse refinement (‘Restricted’ mode) and 0.85 for a final refinement (‘Full’ mode) of the best candidates.
Output
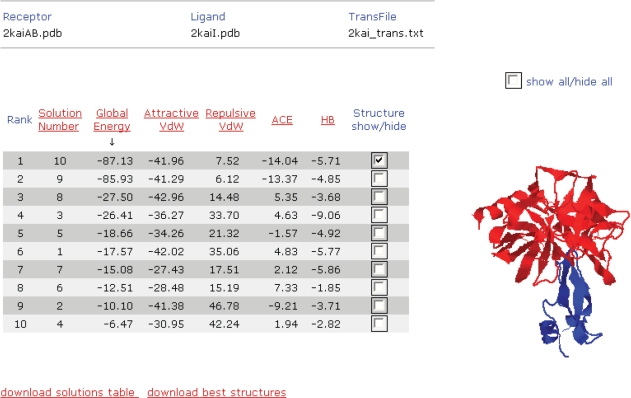
The output of the server is a table of all the input solutions, where each row corresponds to a single input complex (Figure 1). The table is sorted by global energy values. Refined complex structures are generated for up to 100 lowest energy candidates. The user can view the complexes in a Jmol applet window (37). Different complexes can be viewed simultaneously for comparison and the 3D structures can be downloaded as PDB files. The table can be sorted by different energy terms, such as the attractive and repulsive van der Walls forces, the ACE and the contribution of the hydrogen bonds (HB) to the global binding energy. An extended table with full specifications of the values of each energy term can be downloaded.
Figure 1.
The output table of FireDock server.
CONCLUSIONS
The FireDock web server presented here is the first web server for addressing the problem of protein flexibility in docking and allows a high-throughput refinement of docking candidates. Due to the high efficiency of the algorithm, we are able to present a web server that allows online refinement of large number of candidates within reasonable running times. The web server interface is easy to understand and simple, requiring no previous knowledge in docking algorithms. In addition, we provide an option for redirection of the solution candidates from the PatchDock web server to the refinement process of FireDock. This allows the users to perform a full docking protocol of global search (PatchDock) and local refinement (FireDock) in a fully automated manner. The algorithm of FireDock was proven to be successful in the task of refinement and selection of near-native solution out of thousand candidates (16). We believe that the server will enable biologists to use this state-of-the-art docking algorithm in order to predict the structure of new protein–protein complexes.
ACKNOWLEDGEMENTS
We thank Oranit Dror for the helpful comments. E.M. was supported in part by a fellowship from the Edmond J. Safra Bioinformatics Program at Tel-Aviv university. The research of H.J.W has been supported in part by the Israel Science Foundation (grant no. 281/05) and by the Hermann Minkowski-Minerva Center for Geometry at TAU. The research of HJW and RN has also been supported in part by the NIAID, NIH (grant No. 1UC1AI067231) and by the Binational US-Israel Science Foundation (BSF). This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract number NO1-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Funding to pay the Open Access publication charges for this article was provided by NCI contract N01-CO-12400.
Conflict of interest staetement. None declared.
REFERENCES
- 1.Cusick ME, Klitgord N, Vidal M, Hill DE. Interactome: gateway into systems biology. Hum. Mol. Genet. 2005;14:171–181. doi: 10.1093/hmg/ddi335. [DOI] [PubMed] [Google Scholar]
- 2.Grabowski M, Joachimiak A, Otwinowski Z, Minor W. Structural genomics: keeping up with expanding knowledge of the protein universe. Curr. Opin. Struct. Biol. 2007;17:347–353. doi: 10.1016/j.sbi.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aloy P, Russell RB. Structural systems biology: modelling protein interactions. Nat. Rev. Mol. Cell. Bi. 2006;7:188–197. doi: 10.1038/nrm1859. [DOI] [PubMed] [Google Scholar]
- 4.Goh CS, Milburn D, Gerstein M. Conformational changes associated with protein-protein interactions. Curr. Opin. Chem. Biol. 2004;14:1–6. doi: 10.1016/j.sbi.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 5.May A, Zacharias M. Accounting for global protein deformability during protein-protein and protein-ligand docking. Biochim. Biophys. Acta. 2005;1754:225–231. doi: 10.1016/j.bbapap.2005.07.045. [DOI] [PubMed] [Google Scholar]
- 6.Bonvin AM. Flexible protein-protein docking. Curr. Opin. Struct. Biol. 2006;16:194–200. doi: 10.1016/j.sbi.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Halperin I, Ma B, Wolfson HJ, Nussinov R. Principles of docking: an overview of search algorithms and a guide to scoring functions. Proteins. 2002;47:409–442. doi: 10.1002/prot.10115. [DOI] [PubMed] [Google Scholar]
- 8.Schneidman-Duhovny D, Nussinov R, Wolfson HJ. Predicting molecular interactions in silico II: protein-protein and protein-drug docking. Front. Med. Chem. 2006;3:585–613. doi: 10.2174/0929867043456223. [DOI] [PubMed] [Google Scholar]
- 9.Eisenstein M, Katchalski-Katzir E. On proteins, grids, correlations, and docking. C R Biol. 2004;327:409–420. doi: 10.1016/j.crvi.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 10.Duhovny D, Nussinov R, Wolfson HJ. Efficient unbound docking of rigid molecules. In: Guigo R, Gusfield D, editors. Proceedings of the Fourth International Workshop on Algorithms in Bioinformatics. Vol. 2452. Italy: Springer-Verlag GmbH Rome; 2002. pp. 185–200. [Google Scholar]
- 11.Palma PN, Krippahl L, Wampler JE, Moura J. BIGGER: A new (soft) docking algorithm for predicting protein interactions. Proteins. 2000;39:372–384. [PubMed] [Google Scholar]
- 12.Gray J, Moughon S, Schueler-Furman O, Kuhlman B, Rohl C, Baker D. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J. Mol. Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 13.Fernndez-Recio J, Totrov M, Abagyan R. ICM-DISCO docking by global energy optimization with fully exible side-chains. Proteins. 2003;52:113–117. doi: 10.1002/prot.10383. [DOI] [PubMed] [Google Scholar]
- 14.Canutescu AA, Shelenkov A, Dunbrack R.L., Jr A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 2003;12 doi: 10.1110/ps.03154503. 2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu J. Rapid protein side-chain packing via tree decomposition. Lect. Notes Comp. Sci. 2005;3500:423–439. [Google Scholar]
- 16.Andrusier N, Nussinov R, Wolfson HJ. FireDock: fast interaction refinement in molecular docking. Proteins. 2007;69:139–159. doi: 10.1002/prot.21495. [DOI] [PubMed] [Google Scholar]
- 17.Jackson R, Gabb H, Sternberg M. Rapid refinement of protein interfaces incorporating solvation: application to the docking problem. J. Mol. Biol. 1998;276:265–285. doi: 10.1006/jmbi.1997.1519. [DOI] [PubMed] [Google Scholar]
- 18.Li L, Chen R, Weng Z. RDOCK: refinement of rigid-body protein docking predictions. Proteins. 2003;53:693–707. doi: 10.1002/prot.10460. [DOI] [PubMed] [Google Scholar]
- 19.Camacho JC, Vajda S. Protein docking along smooth association pathways. Proc. Natl Acad. Sci. USA. 2001;98:10636–10641. doi: 10.1073/pnas.181147798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen R, Li L, Weng Z. ZDOCK: An initial-stage protein-docking algorithm. Proteins. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 21.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Comeau S, Gatchell D, Vajda S, Camacho C. ClusPro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res. 2004;32:W96–99. doi: 10.1093/nar/gkh354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics. 2004;20:45–50. doi: 10.1093/bioinformatics/btg371. [DOI] [PubMed] [Google Scholar]
- 24.Camacho CJ, Gatchell DW. Successful discrimination of protein interactions. Proteins. 2003;52:92–97. doi: 10.1002/prot.10394. [DOI] [PubMed] [Google Scholar]
- 25.Wang C, Schueler-Furman O, Baker D. Improved side-chain modeling for protein-protein docking. Protein Sci. 2005;14:1328–1339. doi: 10.1110/ps.041222905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tovchigrechko A, Vakser IA. GRAMM-X public web server for protein–protein docking. Nucleic Acids Res. 2006;34:310–314. doi: 10.1093/nar/gkl206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ritchie D, Kemp G. Protein docking using spherical polar fourier correlations. Proteins. 2000;39:178–194. [PubMed] [Google Scholar]
- 28.Chen R, Mintseris J, Janin J, Weng Z. A protein-protein docking benchmark. Proteins. 2003;52:88–91. doi: 10.1002/prot.10390. [DOI] [PubMed] [Google Scholar]
- 29.Mintseris J, Wiehe K, Pierce B, Anderson R, Chen R, Janin J, Weng Z. Protein-protein docking benchmark 2.0: an update. Proteins. 2005;60:214–216. doi: 10.1002/prot.20560. [DOI] [PubMed] [Google Scholar]
- 30.Eriksson O. Side chain-positioning as an integer programming problem. LNCS. 2001;2149:128–141. [Google Scholar]
- 31.Althaus E, Kohlbacher O, Lenhof HP, Muller P. A combinatorial approach to protein docking with exible side chains. J. Comp. Biol. 2002;9:597–612. doi: 10.1089/106652702760277336. [DOI] [PubMed] [Google Scholar]
- 32.Kingsford L, Bernard C, Mona S. Solving and analyzing side-chain positioning problems using linear and integer programming. Bioinformatics. 2005;21:1028–1039. doi: 10.1093/bioinformatics/bti144. [DOI] [PubMed] [Google Scholar]
- 33.Broyden CG. The convergence of a class of double-rank minimization algorithms. J. Inst. Math. Appl. 1970;6:76–90. [Google Scholar]
- 34.Fletcher R. A new approach to variable metric algorithms. Comput. J. 1970;13:317–322. [Google Scholar]
- 35.Berman H, Westbrook J, Feng Z, Gilliland G, Bhat T, Weissig H, Shindyalov I, Bourne P. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen R, Weng Z. Docking unbound proteins using shape complementarity, desolvation, and electrostatics. Proteins. 2002;47:281–294. doi: 10.1002/prot.10092. [DOI] [PubMed] [Google Scholar]
- 37.Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochemistry and Molecular Biology Education. 2006;34:255–261. doi: 10.1002/bmb.2006.494034042644. [DOI] [PubMed] [Google Scholar]