Abstract
Jpred (http://www.compbio.dundee.ac.uk/jpred) is a secondary structure prediction server powered by the Jnet algorithm. Jpred performs over 1000 predictions per week for users in more than 50 countries. The recently updated Jnet algorithm provides a three-state (α-helix, β-strand and coil) prediction of secondary structure at an accuracy of 81.5%. Given either a single protein sequence or a multiple sequence alignment, Jpred derives alignment profiles from which predictions of secondary structure and solvent accessibility are made. The predictions are presented as coloured HTML, plain text, PostScript, PDF and via the Jalview alignment editor to allow flexibility in viewing and applying the data. The new Jpred 3 server includes significant usability improvements that include clearer feedback of the progress or failure of submitted requests. Functional improvements include batch submission of sequences, summary results via email and updates to the search databases. A new software pipeline will enable Jnet/Jpred to continue to be updated in sync with major updates to SCOP and UniProt and so ensures that Jpred 3 will maintain high-accuracy predictions.
INTRODUCTION
Despite recent structural genomics initiatives (1,2), the disparity between knowledge of protein structure and sequence continues to grow larger. Currently, there are less than 50 000 structures stored in the PDB (3), compared to almost 5 million sequences in UniProt Release 12 (4). Prediction of protein structure by homology modelling methods (5,6) is the most reliable approach, but depending on the genome, >50% of proteins lack a structural homologue that is similar enough to the query to build a confident model (7,8). Accordingly, since knowledge of protein structure is key in understanding the detailed function and pathology of a protein there is intense interest in gleaning structural details from sequence alone. Although there have been recent advances in ab initio protein structure prediction (9–12) it is still not possible routinely to predict reliable, detailed protein 3D structures in the absence of a homologue of known structure.
Techniques for secondary structure prediction, the identification of regions of α-helix, β-strand and ‘coil’, go some way to fill this void. Early techniques to predict protein secondary structure worked from a single sequence (13–15). With the growth in available sequences and availability of automatic multiple protein sequence alignment methods (16), Zvelebil et al. (17) first demonstrated that the information from a multiple sequence alignment gave an increase in accuracy of 9% (to 66%) on 11 proteins. The original Jpred server exploited the further finding that combination of several good predictors led to an improvement in accuracy to 72.9% in a blind test on 396 proteins (18). This, and the pioneering work of Rost and Sander in their PHD neural network program (19) led to the development of the Jnet (Joint Network) neural network predictor that combined multiple neural networks, which had been trained on the same multiple sequence alignments, but where the alignments were presented to the networks in different ways. Jnet raised the accuracy of secondary structure prediction to 76.4% in a blind test on 480 proteins (20). Similar accuracy has been achieved by the PSIPRED algorithm, which also predicts from multiple alignment profiles (21). The Jnet algorithm has formed the basis of the Jpred prediction server since 2000, but recent work to optimize Jnet and retrain on extended databases has raised the accuracy of Jnet to 81.5% in blind tests (Cole and Barton, manuscript submitted for publication). Here, we describe significant updates to the Jpred server which now includes the improved Jnet v2.0 algorithm.
METHODS
The Jnet algorithm
The Jpred server takes a single protein sequence or multiple sequence alignment (MSA) and returns predictions made by the Jnet algorithm. The details of Jnet v2.0 will be described elsewhere (Cole and Barton, manuscript submitted for publication) but follow similar lines to Jnet v1.0 (20). The main differences in Jnet v2.0 are the use of only PSI-BLAST (22) Position-specific scoring matrix (PSSM) and HMMER (23) hidden Markov model (HMM) profiles (21) rather than including frequency profiles, and moving from 9 to 100 hidden units in the neural networks. The method was developed through 7-fold cross-validated training on a sequence and structure non-redundant dataset derived from the Astral compendium of SCOP domain data (release 1.71) (24,25) at the superfamily level. Testing on a blind dataset of 149 sequences gave a final secondary structure prediction Q3 score of 81.5%, which is ∼5% better than previously published for Jnet (20). Solvent accessibility accuracy was found to be 88.9%, 82.4% and 77.8% for each of >0%, >5% and >25% relative solvent accessibility thresholds, a 1.5–2.5% improvement over Jnet v1.0.
Protein sequence input
The most reliable way to predict protein secondary structure is by similarity to a protein of known structure. Accordingly, user submitted sequences are first searched with BLAST (26) against sequences in PDB (3) (0.0005 e-value cut-off). The user can choose to skip this step, or can continue to the prediction from the PDB hit output (if any). The PDB search step is only used to inform the user that a similar protein with known structure exists and is not used to further inform the secondary structure prediction like other methods (27,28). The protein sequence is then searched against UniRef90 (29) and an alignment constructed with PSI-BLAST (22) (three iterations, a first iteration e-value cut-off of 0.05 and 0.01 e-value cut-off for subsequent iterations). All alignments are filtered for redundancy at 75% sequence identity since this was found to be optimal in earlier work (20). An HMM profile is obtained from the alignment via the HMMer program (23) and a PSSM profile is output by PSI-BLAST. The two profiles are input into Jnet for predicting secondary structure and solvent accessibility.
It can be time consuming to search UniRef90 for hits to the query and to prepare the inputs for Jnet, so to aid overall throughput, jobs are aborted if they exceed 1 h of CPU time. The user is informed interactively and/or via email, if an address has been provided. There is no time limit for queuing jobs.
Once the job is finished, a results page is generated with links to HTML, postscript and PDF outputs. The outputs include the alignment generated by Jpred, Jnet predictions of solvent accessibility at >0%, >5% and >25% relative accessibility cut-offs, Jnet secondary structure prediction (including the predictions made by the separate PSSM and HMM networks) and coiled-coil predictions performed by the multicoil application (30). Jnet, also, assigns a confidence score (low, 0 to high, 9) to each predicted residue's secondary structure which can be viewed in the Jpred output.
An interactive output of the results is available through the Jalview alignment viewer applet (31), allowing the user to edit the alignment as required.
Results are only stored on the server for 2 days, but all the data are available for download for local storage.
In supplying an email address, users need not follow submissions interactively, but may wait to receive an email when the submission has completed.
MSA Input
Jpred can also accept a user-generated MSA as input for secondary structure prediction in FASTA, MSF or BLC format. If an MSA is provided, searching against the PDB for structural homologues and UniRef90 is not performed and so a PSI-BLAST PSSM profile is not generated. Accordingly, the secondary structure and solvent accessibility predictions are performed from only the HMM profile obtained in passing the alignment through HMMer. Excluding the PSI-BLAST PSSM may reduce the accuracy of the resulting prediction. For example, in a blind test on 149 proteins using PSI-BLAST generated alignments, the prediction accuracy drops from 81.5% to 80.3% on average when only HMMer profiles are input. However, since the user-supplied alignment might not share the same characteristics as a PSI-BLAST alignment it is difficult to assess accurately the impact of not including the PSSM. One benefit of supplying an alignment is that predictions are typically returned in <2 min, since no time-consuming database search is performed.
Improvements and Additions
Version 3 of the Jpred server has been completely overhauled with many visible and invisible changes over Jpred 2 (20). The main visible changes are listed below.
User interface
From a usability perspective, the user interface has been updated and is now fully XHTML 1.0 and CSS 2.0 compliant as validated by the World Wide Web Consortium validation service (http://validator.w3.org). This ensures that the server will work equivalently in all compliant web browsers.
Submission of a sequence has been simplified for first-time users by including a straight-forward text box on the homepage. Giving a raw sequence and clicking ‘Make Prediction’ submits an interactive job, which the user follows until completion. This includes checking the PDB for similar sequences. The ‘Advanced’ link permits users familiar with Jpred to access options allowing more control over their submissions. Advanced options include choice of input format, provision of user supplied job name, email address and a toggle of whether a search of the PDB is required. Allowing users to supply their own job name is a new feature following feedback from users. If the user also supplies an email address it then makes collating results from several jobs much easier than having randomly generated Jpred job names.
On pressing the ‘Make Prediction’ button the input sequence or alignment is thoroughly checked as being valid input and any errors are reported to the user with suggestions on how to rectify the problem. The sequence is checked against the PDB at this stage and any hits reported. As shown in Figure 1, it is possible now to click through to the prediction rather than having to re-submit requests which have hits to the PDB. Valid requests are added to the Jpred queue waiting to be run. Once a job request is running the new progress meter, which updates every 10 s as illustrated in Figure 1, gives an easy indication of how the submission is proceeding in increments of 10% completion. A link to the raw text output of Jpred's progress is also present on the progress page. If an email address is supplied, there is no need to keep the browser window open as an email will be sent informing that the job is complete and ready for viewing.
Figure 1.
Examples of progress of a submission to Jpred. A screenshot of a list of hits to the PDB (background) and of the Jpred progress meter (foreground). The ‘continue’ button on the PDB hits page allows the user to continue on to the submission of a secondary structure prediction job.
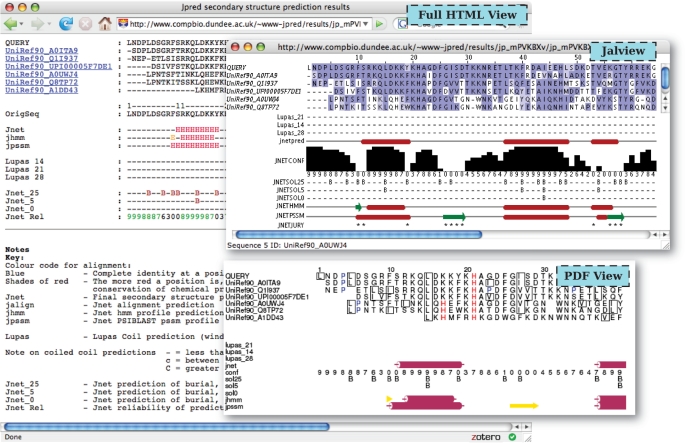
When the Jpred submission has completed, the user is re-directed to the results page, which includes links to several viewing options. The layout of the results page has been improved making it less verbose and clearer to use. The ‘Simple HTML’ output shows an alignment of the query sequence with the prediction, colour-coded by secondary structure type (helix is red, sheet is yellow and coil is black) and the ‘Full HTML’ shows the prediction in the context of the full MSA generated by PSI-BLAST or provided by the user. Solvent accessibility predictions are shown also in the ‘Full HTML’ output. A printable version of the ‘Full HTML’ output is presented as postscript and portable document format (PDF) files created by the ALSCRIPT application (32). The alignment and prediction data can additionally be viewed in the Jalview interactive alignment viewer applet (31). Examples of the Full HTML, PDF and Jalview outputs are shown in Figure 2.
Figure 2.
Jpred results viewing options. The ‘Full HTML’, ‘PDF’ and ‘Jalview’ views of the results are shown for the same query sequence and the different representations of the secondary structure, solvent accessibility and coiled-coil predictions can be compared.
Results emails
A simple output of the secondary structure prediction is sent via email to the user-supplied address together with the query name (in the subject header) and sequence. For more detailed information regarding the prediction or for solvent accessibility data, a URL is provided in the email to the Jpred results page.
Batch submission
Batch submission of job requests is a new feature of Jpred 3. Batch submissions are limited to a maximum of 20 sequences per submission and are required in FASTA format. Each sequence in the FASTA file is treated as a single job request, but without searching for similarities in the PDB. Sequence names are taken from the FASTA description line and used as the job name. As batch submissions cannot be viewed interactively, an email address is required upon submission and an email is returned for each sequence in the batch submission as for a standard submission to Jpred.
Orphan proteins
Occasionally, query sequences have no hits to UniRef90 and previously Jpred would have failed on these sequences. In these instances, Jpred 3 makes a prediction based only on an HMM profile created from the query sequence. However, average prediction accuracy is significantly worse than when an alignment is available. Blind testing on 149 sequences shows a drop in accuracy from 81.5% with full alignment data to 65.9% with only single sequence data, highlighting the importance of having alignment data. Although the prediction accuracy from a single sequence is low it is still significantly better than random (∼42%).
Updates
The PDB sequence database that is searched for similarities is updated weekly. Secondary structure prediction performs best when it has up-to-date data for constructing alignments as input. The UniRef90 search database for constructing alignments has been updated to version 10.1 and will be updated at least as often as Jnet is retrained. In addition to good alignments, larger and non-redundant training datasets have also been shown to be beneficial for improved prediction accuracies (Cole and Barton, manuscript submitted for publication) (33). Hence, Jnet will now be linked to SCOP releases and will be retrained whenever a new SCOP release is announced or soon thereafter. The datasets used for training Jnet will be made available via the website.
A new semi-automatic pipeline has been developed in Perl for creating the Jnet training data. The pipeline requires SCOP domain definition and sequence data as determined by Astral and will create (with some manual input) a structurally and sequence non-redundant dataset ready for input to Jnet neural networks for full-scale training and validation via the SNNS application. Once the training of Jnet is checked on an independent blind set it will be recompiled with the new networks and used in Jpred.
CONCLUSIONS
Secondary structure prediction is an important tool in a structural biologist's toolbox for the analysis of the significant numbers of proteins, which have no sequence similarity to proteins of known structure. Jpred is a secondary structure prediction server that is a well used and accurate source of predicted secondary structure.
The recent update of Jpred incorporates the latest version of the Jnet algorithm improving secondary structure prediction to 81.5% and solvent accessibility predictions to up to 88.9%. The most obvious changes to the server are to do with user interaction with Jpred by giving more feedback to users regarding problems or progress of their submissions. Including better checking of input data and a progress meter during running jobs allows for more successful results. The variety of results viewing options gives users flexibility in how they wish to present their data.
More fundamentally, submissions can now be made in a batch-wise manner and secondary structure predictions are returned via email if an address is supplied. In order to keep Jpred up-to-date with new sequence information, a pipeline has been developed to retrain the Jnet algorithm and update all the relevant databases on a regular basis.
ACKNOWLEDGEMENTS
We thank Drs Tom Walsh and Jonathan Monk for computational support, and Dr Jim Procter and Jonathan Manning for helpful comments. Funding was provided by the Scottish Funding Council (SFC) as part of the Scottish Bioinformatics Research Network (SBRN) initiative. Funding to pay the Open Access publication charges for this article was provided by the SFC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Chen L, Oughtred R, Berman HM, Westbrook J. TargetDB: a target registration database for structural genomics projects. Bioinformatics. 2004;20:2860–2862. doi: 10.1093/bioinformatics/bth300. [DOI] [PubMed] [Google Scholar]
- 2.Norvell JC, Machalek AZ. Structural genomics programs at the US National Institute of General Medical Sciences. Nat. Struct. Biol. 2000;7(Suppl):931. doi: 10.1038/80694. [DOI] [PubMed] [Google Scholar]
- 3.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The UniProt Consortium. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2007;35:D193–D197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 6.Dalton JA, Jackson RM. An evaluation of automated homology modelling methods at low target template sequence similarity. Bioinformatics. 2007;23:1901–1908. doi: 10.1093/bioinformatics/btm262. [DOI] [PubMed] [Google Scholar]
- 7.Chandonia JM, Brenner SE. Implications of structural genomics target selection strategies: Pfam5000, whole genome, and random approaches. Proteins. 2005;58:166–179. doi: 10.1002/prot.20298. [DOI] [PubMed] [Google Scholar]
- 8.Marsden RL, Lewis TA, Orengo CA. Towards a comprehensive structural coverage of completed genomes: a structural genomics viewpoint. BMC Bioinform. 2007;8:86. doi: 10.1186/1471-2105-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradley P, Misura KM, Baker D. Toward high-resolution de novo structure prediction for small proteins. Science. 2005;309:1868–1871. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- 10.Jones DT, Bryson K, Coleman A, McGuffin LJ, Sadowski MI, Sodhi JS, Ward JJ. Prediction of novel and analogous folds using fragment assembly and fold recognition. Proteins. 2005;61(Suppl 7):143–151. doi: 10.1002/prot.20731. [DOI] [PubMed] [Google Scholar]
- 11.Qian B, Raman S, Das R, Bradley P, McCoy AJ, Read RJ, Baker D. High-resolution structure prediction and the crystallographic phase problem. Nature. 2007;450:259–264. doi: 10.1038/nature06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones DT. Predicting novel protein folds by using FRAGFOLD. Proteins. 2001;45 (Suppl 5):127–132. doi: 10.1002/prot.1171. [DOI] [PubMed] [Google Scholar]
- 13.Chou PY, Fasman GD. Prediction of the secondary structure of proteins from their amino acid sequence. Adv. Enzymol. Relat. Areas Mol. Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 14.Garnier J, Osguthorpe DJ, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J. Mol. Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 15.Lim VI. Algorithms for prediction of alpha-helical and beta-structural regions in globular proteins. J. Mol. Biol. 1974;88:873–894. doi: 10.1016/0022-2836(74)90405-7. [DOI] [PubMed] [Google Scholar]
- 16.Barton GJ, Sternberg MJ. Evaluation and improvements in the automatic alignment of protein sequences. Protein Eng. 1987;1:89–94. doi: 10.1093/protein/1.2.89. [DOI] [PubMed] [Google Scholar]
- 17.Zvelebil MJ, Barton GJ, Taylor WR, Sternberg MJ. Prediction of protein secondary structure and active sites using the alignment of homologous sequences. J. Mol. Biol. 1987;195:957–961. doi: 10.1016/0022-2836(87)90501-8. [DOI] [PubMed] [Google Scholar]
- 18.Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- 19.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 20.Cuff JA, Barton GJ. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 21.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 22.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 24.Brenner SE, Koehl P, Levitt M. The ASTRAL compendium for protein structure and sequence analysis. Nucleic Acids Res. 2000;28:254–256. doi: 10.1093/nar/28.1.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 26.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Montgomerie S, Sundararaj S, Gallin WJ, Wishart DS. Improving the accuracy of protein secondary structure prediction using structural alignment. BMC Bioinform. 2006;7:301. doi: 10.1186/1471-2105-7-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pollastri G, Martin AJ, Mooney C, Vullo A. Accurate prediction of protein secondary structure and solvent accessibility by consensus combiners of sequence and structure information. BMC Bioinform. 2007;8:201. doi: 10.1186/1471-2105-8-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. UniRef: comprehensive and non-Redundant UniProt reference clusters. Bioinformatics. 2007;23:1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 30.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 31.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 32.Barton GJ. ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- 33.Rost B. Review: protein secondary structure prediction continues to rise. J. Struct. Biol. 2001;134:204–218. doi: 10.1006/jsbi.2001.4336. [DOI] [PubMed] [Google Scholar]