Abstract
A particular challenge in biomedical text mining is to find ways of handling ‘comprehensive’ or ‘associative’ queries such as ‘Find all genes associated with breast cancer’. Given that many queries in genomics, proteomics or metabolomics involve these kind of comprehensive searches we believe that a web-based tool that could support these searches would be quite useful. In response to this need, we have developed the PolySearch web server. PolySearch supports >50 different classes of queries against nearly a dozen different types of text, scientific abstract or bioinformatic databases. The typical query supported by PolySearch is ‘Given X, find all Y's’ where X or Y can be diseases, tissues, cell compartments, gene/protein names, SNPs, mutations, drugs and metabolites. PolySearch also exploits a variety of techniques in text mining and information retrieval to identify, highlight and rank informative abstracts, paragraphs or sentences. PolySearch's performance has been assessed in tasks such as gene synonym identification, protein–protein interaction identification and disease gene identification using a variety of manually assembled ‘gold standard’ text corpuses. Its f-measure on these tasks is 88, 81 and 79%, respectively. These values are between 5 and 50% better than other published tools. The server is freely available at http://wishart.biology.ualberta.ca/polysearch
INTRODUCTION
Today's scientists are deluged with information. Currently there are >8000 scientific, technical and medical journals publishing >1 000 000 articles a year. Nearly 40% of these articles are biomedical in nature. Indeed, it has been estimated that in order for a scientist to stay current for a single high-priority disease (say breast cancer), they would have to scan 130 different journals and read 27 papers each week (1). Given that most journal articles are not exactly ‘light’ reading, this task of staying current with the literature could easily occupy 75% of a scientist's working day. The problem with information overload is not restricted to scientific papers. Electronic databases are equally culpable. Thousands of web accessible text, image and sequence databases now exist (2). These contain terabytes of data and are expanding in both number and size far faster than the rate of scientific publishing. Just tracking the appearance and content of new databases, let alone using the information in them, can prove to be a full-time challenge.
Clearly, the quantity of information generated by the scientific community is far too great for any human to efficiently process or assimilate. Too much fragmentary information and non-contextual data exists in too many places. This makes the task of finding relevant information on a specialized topic somewhat like finding a needle in the proverbial haystack. It is now obvious that a key challenge, especially in the field of bioinformatics, is to develop methods that allow this information to be easily retrieved and readily exploited by human users. One route is to pre-compute or synopsize this information and assemble it into specialized biomedical databases or encyclopedias. However, given the need for constant updating, expert manual curation and the wide variety of biomedical topics that users may want to access, pre-assembled databases are not a perfect panacea to the problem of information overload. An alternative route is to improve the tools for automated or semi-automated biomedical information retrieval.
An important advance in biomedical information retrieval has come with the development of NCBI's Entrez Cross-Database search system (3). This system brings the hunt for new and useful biomedical data to a new level by integrating PubMed (i.e. biomedical abstract data) with NCBI's multitude of sequence, structure and chemical databases. Entrez is a superb resource that greatly improves the speed and precision with which researchers can find relevant data on a given gene, disease, mutation, drug or microarray experiment.
However, Entrez is still somewhat limited because it is restricted to searching its abstract and molecular database resources only through MeSH (Medical Subject Heading) terms, MeSA (Medical Subject Annotation) terms and keywords in database titles or database names. In other words, Entrez is not capable of scanning through the full text of all 168 000 abstracts on, say, breast cancer assembling a list of genes that are mentioned in those abstracts, extracting key sentences for those genes, counting the frequency of appearance of those genes and providing a frequency or relevancy ranking for them. Likewise, Entrez does not link its results to many equally useful external databases such as SwissProt (4), the Human Gene Mutation Database (HGMD) (5), DrugBank (6) or the Human Metabolome Database (HMDB) (7). Another unfortunate limitation is that Entrez does not contain disease, gene/protein, drug or metabolite compendia (i.e. all entity lists). For instance, if one wanted to find all the drugs that could be used to treat breast cancer, one would have to repeatedly enter ‘breast cancer AND Y’ where Y is the name of each of the 25 000 known drug brands.
These kind of sophisticated text searching tasks are more suited to a different class of programs called medical text mining systems. Several excellent web-based biomedical text mining tools now exist such as MedMiner (8), MedGene (9), LitMiner (10), iHOP (11), ALIBABA (12) and EBIMed (13). These tools exploit the explicit textual information contained within the PubMed database by selecting or highlighting key sentences or terms within the abstracts and then presenting the results in some form. However, these text mining tools were designed specifically to extract information only from PubMed abstracts and not other databases (i.e. SwissProt, HGMD and DrugBank). Ideally what is needed is something that combines the text mining capabilities found in MedMinder or EBIMed with the database integration found in Entrez. What's more, one would like to see analytical capabilities built into such a system so that users could manipulate, view, or archive the resulting information (text or sequence) in a convenient, web accessible format. These requirements motivated us to develop just such a resource—called PolySearch.
PolySearch is a web accessible tool that is designed specifically for extracting and analyzing text-derived relationships between human diseases, genes/proteins, mutations (SNPs), drugs, metabolites, pathways, tissues, organs and sub-cellular localizations. It also displays links and ranks text, as well as sequence data in multiple forms and formats. A distinguishing feature of PolySearch over other biomedical text mining tools is the fact that it extracts and analyses not only PubMed data, but also text data from multiple databases (DrugBank, SwissProt, HGMD, Entrez SNP, etc.). This integration of current literature text and database ‘factoids’ allows PolySearch to extract and rank information that is not easily found in databases alone or in journals alone. A more detailed description of PolySearch follows.
Implementation
PolySearch, as the name suggests, is a tool that supports multiple (‘poly’) types of biomedical text searches from multiple (‘poly’) types of databases. It is also designed to facilitate the search, retrieval and compilation of disease-associated human ‘poly’morphisms (SNPs). PolySearch exploits recent advances in text mining along with the readily availability of diverse biomedical databases and biomedical thesauruses to permit a wide variety of complex or expansive text searches over many biomedical domains. PolySearch consists of seven basic components: (i) a web-based user interface for constructing queries; (ii) a collection of internal and external biomedical databases; (iii) a collection of biomedical synonyms (custom thesauruses and all entity lists); (iv) a general text search engine for extracting data from heterogeneous databases; (v) a schema for selecting, ranking and integrating content; (vi) a display tool for displaying and synopsizing results and (vii) a PCR primer-designing tool to facilitate SNP and mutation studies. A figure outlining PolySearch's general design and the databases it uses is given in the PolySearch ‘Documentation’ page.
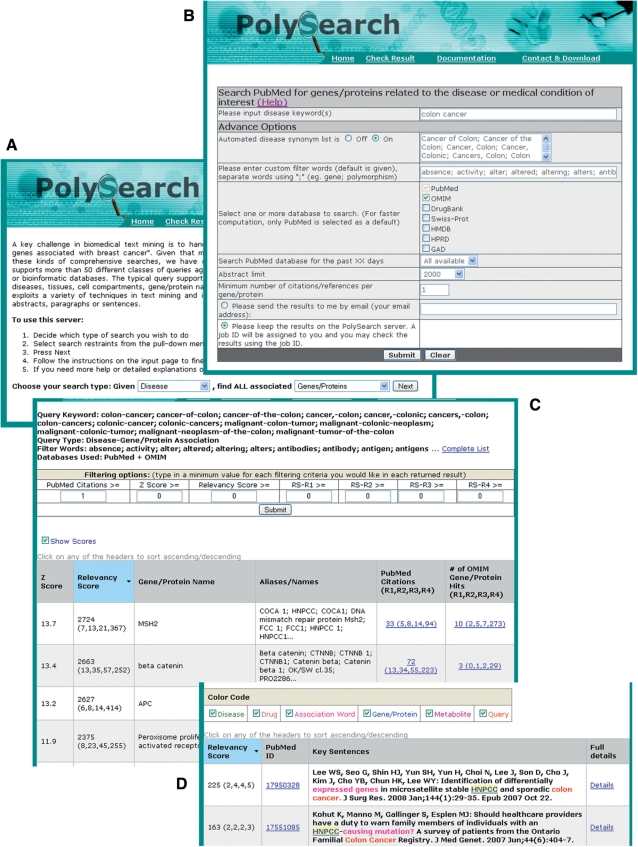
PolySearch's query interface was written in standard HTML and Perl. PolySearch has been tested on a variety of platforms is compatible with most common browsers (Netscape, Firefox, Safari and Internet Explorer). It uses a series of text boxes and pull-down menus to facilitate query construction. A screen shot of the query interface is shown in Figure 1A. The basic structure of almost every PolySearch query is ‘given a single X find all associated Y's’, where X can be any single human disease, gene/protein name, drug, metabolite, SNP, gene/protein sequence or user-provided text word and Y can be any one of all human diseases, genes/proteins, drugs, metabolites, pathways, tissues, organs, sub-cellular localizations, SNPs, PCR primers or user-supplied text words. In each case the ‘X’ and ‘Y’ words can correspond to either a common name or synonyms. For future reference we will refer to ‘X’ as the ‘query term’ and ‘Y’ as the ‘database term’. Table 1 provides a more detailed listing of all allowed queries in PolySearch. Once the general query is constructed and submitted the user is presented with a second page (the query refinement page—Figure 1B) that allows further refinement of the query, including the selection of association words, databases, query word synonyms and display options.
Figure 1.
A screenshot montage of PolySearch's query interface and result display showing: (A) the PolySearch query interface; (B) the query refinement page; (C) the PolySearch result table where both PubMed and OMIM were searched and (D) the sentence and keyword display view obtained by clicking on the PubMed citation links in the result table.
Table 1.
A detailed listing of all allowed ‘basic’ queries in PolySearch
| Given | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Disease | Gene/protein | Drug | Metabolite | Text word | Pathway | Tissue | SNP (RS#) | Gene/protein sequence | |
| Find | |||||||||
| Disease | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Gene/protein | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Drug | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Metabolite | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Tissue | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Organ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Subcellular localization | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Pathway | ✓ | ✓ | ✓ | ✓ | ✓ | ||||
| Text word | ✓ | ||||||||
| SNP | ✓ | ✓ | |||||||
| PCR primers | ✓ | ✓ | |||||||
Through its query refinement page, PolySearch also allows users to add or include synonyms to their original query words (i.e. query synonym expansion). In particular, PolySearch uses its own thesauruses to automatically append synonyms to a query word (by clicking on the option for ‘automated synonym list’). If the computer-generated synonyms appear inadequate, the user may further edit or add to this list. Users can also edit the set of association words used to refine PolySearch queries. Association words play a key role in improving the precision and recall of any given PolySearch query. For each of the 50+ query combinations available through PolySearch, sets of association words (ranging in number from 40 to 400) were developed through extensive manual testing and iterative modification. Complete list of association words are available in the PolySearch Documentation web pages.
From the query refinement interface users can also choose to limit their search to PubMed only, or to perform their search on some of PolySearch's other reference databases. Limiting PolySearch searches to the PubMed database (the default configuration) is faster but the results tend to be less accurate. Additionally, through the query refinement interface users can also specify: (i) how far back in time the PubMed records should be searched, (ii) the number of abstracts to be searched and (iii) the minimum number of PubMed citations required to be considered as a hit. Changing these values judiciously can also shorten the search times.
After submitting the query, a progress bar appears indicating the expected length of time that the query would take. PolySearch caches all its queries so that for the more common queries the results can be returned almost instantly. However, for more unique queries the typical time taken for text processing is 1–2 min (PolySearch processes ∼10 abstracts per second). Depending on the server load and query type, some jobs may take 15–20 min. Upon completion of the search task PolySearch displays its results in a hyperlinked HTML table (Figure 1C). Given that a typical PolySearch query is ‘Given X, find all Y's’, what is returned is a table that lists, in rank order of relevance, the most probable ‘Y's’ that match the given ‘X’.
As seen in Figure 1C, the top of the table typically summarizes the query word(s), the type of search, the association words used, and the databases used to construct the search. Below this is a display-filtering tool that allows users to select cutoff values related to the minimum number of citations, Z-scores and/or relevancy scores needed for a result to be displayed on the results table. Additional details about the scoring system are provided later. Below the filtering options table is the PolySearch results table. The first column in this table gives the ranking of the database term found by PolySearch (by Z-score), the second column lists each hit's Z-score (the number of standard deviations that the relevancy score is above the mean), the third column gives the total relevancy score for the match term (with a category breakdown), the fourth column displays the database term itself, the fifth column shows the database term's synonyms or aliases, the sixth column displays the hyperlinks to the abstracts or databases for which PolySearch found relevant sentences or phrases. Matches to additional databases (OMIM, HMDB, DrugBank, etc.) are displayed in appended columns. The table may be sorted in ascending or descending order by clicking on the arrows in the column headers. Figure 1C shows a PolySearch result where both PubMed and OMIM (14) were searched.
Clicking the links under the PubMed column (or other database columns) generates a second HTML table that displays the key sentences found in each database abstract or database field along with hyperlinks to the full database record (Figure 1D). As seen in this figure, the extracted sentences are colour coded to facilitate rapid visual scanning (although this colouring can also be selectively turned off). Words marked in red correspond to the query term(s), blue to human genes, green to diseases, brown to drugs, magenta to metabolites and fuchsia to association words (dark yellow is reserved for the other word types such as pathway, tissue, organ, sub-cellular localization and user provided text words). If a query word happens to be a gene, drug, disease or metabolite, the red colour of the query word takes precedence. Words highlighted with a light yellow background are the current thesaurus words that the user is viewing. This highlighting is used to facilitate rapid visual cueing of the association between the query word and the thesaurus word. The same colour-coding scheme is used in PolySearch's fully annotated abstract view. Mousing over the coloured terms will give database links or database accession number associated with these terms, while clicking on a coloured term will launch a web page corresponding to that term's database page. Note that somewhat different tables and views are generated for PolySearch's SNP and primer design queries. These are described in more detail on PolySearch's Documentation web pages.
Databases and algorithms
PolySearch is specifically designed to address biomedical queries of the form ‘Given a single X, find all Y's’, where X and Y are biomedical terms pertaining to human health and biology. In performing this type of search PolySearch initially generates a list of all possible Y's (plus their synonyms or abbreviations) along with all of X's possible synonyms and abbreviations. If the search is being done through PubMed, a formal PubMed query is constructed that uses these terms (along with the appropriate Boolean operators) and the query is submitted via NCBI's E-utilities application programming interface (API). This API allows PubMed abstracts to be batch downloaded from the PubMed website. PolySearch then scans through these abstracts sentence-by-sentence to look for informative sentences. These sentences are scored, grouped in categories, ranked, colour coded and then displayed as described earlier. A similar sentence-by-sentence scanning and scoring process is done when searches are done on any of PolySearch's local databases.
PolySearch does not use part-of-speech tagging, but rather it uses a dictionary or ‘bag-of-words’ approach to identify relevant text associations. Key to the success of dictionary-based text mining is having a comprehensive collection of words and synonyms, all of which are properly normalized or mapped to appropriate database accession numbers. PolySearch maintains nine different thesauruses, compendia or synonym lists for human genes, human proteins, human diseases, approved drugs, endogenous metabolites, protein/gene pathways, human tissues, human organs and sub-cellular localizations. These thesauruses or compendia are obviously critical for many of the expansive queries (‘given one, find many’) supported by PolySearch. They are also critical for providing the sensitivity and specificity for many single-word queries (i.e. the automated synonym feature in the query refinement page).
PolySearch's human gene/protein thesaurus was compiled (and is updated) from the latest releases of SwissProt (4), Entrez Gene (3), the Human Genome Organisation Gene Nomenclature Committee (HGNC) (15) and the Human Protein Reference Database (HPRD) (16). This thesaurus, which had to be extensively hand edited, includes gene and protein names, gene symbols, gene/protein abbreviations, protein complexes (microtubules, ribosomes, etc.) as well as their known synonyms. PolySearch's disease thesaurus was derived from the Unified Medical Language System (UMLS) (17) which was further supplemented with disease terms obtained from OMIM and extensive manual curation. PolySearch's drug thesaurus consists of a list of drug names and synonyms from DrugBank's list of FDA-approved drugs, while its metabolite thesaurus consists of a list of metabolite names and synonyms from all entries in the HMDB. The pathway thesaurus was created using KEGG (18), BioCarta (http://www.biocarta.com/) and other pathway resources followed by extensive manual editing. To create the tissue and organ thesauruses, the tissue and organ list from LitMiner was first combined with a tissue and organ list manually derived from the tissue specificity field in SwissProt. Finally, the sub-cellular localization thesaurus was created from the list of all possible sub-cellular localizations listed in the HPRD. These thesauruses may be downloaded via PolySearch's Download page.
In addition to these synonym collections, PolySearch also maintains local copies of a number of comprehensively annotated databases including SwissProt, HPRD, HMDB, OMIM, DrugBank, the Genetic Association Database (GAD—19) and the Human Gene Mutation Database (HGMD—5). By keeping local copies of these databases it is possible to greatly accelerate the search times. As shown in the Testing and Assessment section, the use of these databases in a PolySearch query substantially improves the quality of the results. These local databases are also used to normalize (i.e. map text terms to database IDs) many of the terms in PolySearch's thesauruses. Because of the size and difficulties associated with daily maintenance, all queries involving PubMed abstracts or SNP retrieval are performed over the web using the respective APIs for PubMed as well as multiple SNP databases including CGAP (20), EntrezSNP (3) and HapMap (21).
A central premise to PolySearch's search strategy is the assumption that the greater the frequency with which an X and Y association occurs within a collection of abstracts or databases, the more significant the association is likely to be. For instance, if COX2 is mentioned in PubMed as being associated with colon cancer 510 times but thioredoxin is associated with colon cancer only once, then one is more likely to have more confidence in the COX2-colon cancer association. Frequency alone is not always the best way to rate a paper or a website for its relevancy. Therefore, in addition to counting the frequency of apparent associations, PolySearch employs a text ranking scheme to score the most relevant sentences and abstracts that associate both the query and match terms with each other.
Specifically, PolySearch tries to find query terms, association words and database terms in order to identify and enumerate what we call R1, R2, R3 and R4 sentences (R stands for relevancy). An R4 sentence is a sentence that contains just one of the database terms and is used only for statistical normalization. An R3 sentence is a sentence that has one of the database terms as well as the query word. An R2 sentence is a sentence that has one of the database terms, one of the query terms, as well as at least one association word. An R1 sentence is the same as an R2 sentence but in addition, an R1 sentence has to pass PolySearch's pattern recognition criteria. PolySearch's pattern recognition system is rule based and details regarding these rules are provided in the PolySearch Documentation web pages. This kind of pattern recognition has been used in other text mining systems (such as ALIBABA) to extract protein–protein interactions (12,22,23). Collectively, we call the R1, R2, R3 and R4 sentence counts the PolySearch Relevancy Index (PRI). For the purposes of generating a quantitative PRI score and calculating Z-scores, R1 sentences are given a value of 50, R2 sentences = 25, R3 sentences = 5 and R4 sentences = 1. The PRI score is the sum of the R1, R2, R3 and R4 sentences. These weights were determined through extensive testing on hundreds of different queries.
Testing and assessment
A text mining tool is only useful if it gives accurate results and extensive coverage in less time than what could be performed using alternative (i.e. non-computational) or competing computational methods. To evaluate PolySearch's performance, we used several different tests or methods. These included (i) a comparison of features and capabilities between PolySearch and other biomedical text mining tools; (ii) a comparative evaluation of gene synonym identification; (iii) a comparative evaluation of PolySearch's ability to identify protein–protein interactions; (iv) an evaluation of PolySearch's ability to identify disease/gene associations; (v) an evaluation of PolySearch's ability to identify drug/drug-target associations; (vi) an evaluation of Polysearch's ability to identify metabolite/enzyme associations and (vii) several real-life assessments relating to its capacity to facilitate or accelerate database annotations.
Space limitations prevent us from providing a complete summary of all of these evaluations or of the statistical methods used to measure the performance. In particular, details on the statistical methods, the results for evaluations #1, #5, #6 and #7, as well as additional details relating to evaluations #2 through #6 (including precision, recall, true/false positives and true/false negatives) are all available at PolySearch's Documentation web page. Furthermore, all of the testing corpuses or ‘gold standard’ datasets used in these evaluations are available in PolySearch's download pages. Here we will only provide a brief overview of some of the results.
For evaluation #2, PolySearch's ability to identify genes and protein names within different sentences or abstracts was assessed with a dataset that the developers of IHOP used in evaluating gene synonym identification for human genes (11). This dataset contains 181 sentences from various PubMed abstracts with an average of ∼2–3 gene names per sentence (the names include symbols, standard names, abbreviations and synonyms). We manually identified all the gene and protein names or abbreviations from the dataset and used this collection as our gold standard to compare to PolySearch's gene synonym identification for the dataset. For this task the precision for PolySearch was 90.1, while for IHOP it was 87.1. The recall for PolySearch was 85.3, while the recall for IHOP was 81.8. Finally, the f-measure (a combined measure of recall and precision) for PolySearch was 87.6 while for IHOP it was 84.4. These results clearly show that PolySearch's performance on gene/protein identification is comparable to that of IHOP.
In evaluation #3, we compared the performance of PolySearch on protein–protein interaction extraction (for five human proteins) against two other text mining systems (EBIMed and IHOP), and a manually curated database (HPRD) that covers protein–protein interactions. The five proteins were WNT6, PITPNM2, KIF5C, SNX2 and DEDD. The complete set of known protein–protein interactions for these proteins was determined through extensive reading of full-text papers and careful review of interaction database compilations by two experts with degrees in biochemistry and bioinformatics. Our ‘gold standard’ results indicated that WNT6 had 20 interacting partners, PITPNM2 had 6 interacting partners, KIF5C had 10 interacting partners, SNX2 had 23 interacting partners and DEDD had 40 interacting partners. All programs were run using their default parameters for protein–protein interaction searching. After evaluating the results we found that the f-measure for PolySearch alone was 69.2, the f-measure for PolySearch with its HPRD option turned on was 80.8, the f-measure for IHOP was 43.6, the f-measure for EBIMed was 25.3 and for HPRD it was 47.7. These data clearly show that PolySearch achieves the highest f-measure, by a significant margin, among the four different tools.
In evaluation #4 we compared the performance of PolySearch, LitMiner, EBIMed and GAD in identifying gene-disease associations for 10 different human diseases (alkaptonuria, cylindromatosis, Gilbert syndrome, McLeod syndrome, motor neuron disease, omphalocele, onchocerciasis, orofacial cleft, synpolydactyly and vitelliform macular dystrophy). The complete set of known gene-disease associations was determined through extensive reading of full-text papers and careful review of gene-disease database compilations by an expert with degrees in biochemistry and bioinformatics. The list of ‘gold-standard’ gene-disease associations is given in the PolySearch download pages. All programs were run using their default parameters. Our results showed that the f-measure for PolySearch alone was 70.2, the f-measure for PolySearch with its GAD and OMIM options turned on was 78.5, the f-measure for EBIMed was 66.0, the f-measure for LitMiner was 5.8 and for GAD it was 27.5. As with the previous tests, these data clearly show that PolySearch, with its database features turned on, performs very well.
CONCLUSION
PolySearch brings a number of useful innovations to the area of biomedical text mining and information retrieval. These include: (i) a diverse and extensive set of category-specific biomedical thesauruses; (ii) the integration of many well-annotated databases (OMIM, DrugBank, SwissProt, HMDB, HPRD and GAD) as supplementary text resources; (iii) a multi-tiered, informative scoring system and (iv) customizable control over how to rank, view and assess text-derived associations. In addition to these innovations, PolySearch also borrows a number of excellent ideas from existing text mining systems, including colour-coded word highlighting schemes, key sentence display, extensive use of hyperlinks and multi-database connectivity.
PolySearch is not without some limitations. As a text mining tool, PolySearch uses a relatively simple dictionary approach to identify biological or biomedical associations. This means PolySearch cannot identify novel or newly named diseases, genes, cell types, drugs or metabolites. Another limitation lies in its inability to extract context or meaning from sentences or terms. Methods that use artificial intelligence (AI), word context or machine learning (ML) methods could potentially improve the current term identification system. Efforts are underway to incorporate these improvements in future releases of PolySearch.
ACKNOWLEDGEMENTS
We would like to thank Haiyan Zhang and Chunyan Meng for their contributions to this work. Funding for this project was provided by the Protein Engineering Network of Centres of Excellence (PENCE), The Alberta Cancer Foundation, NSERC and Genome Prairie (a division of Genome Canada). Funding to pay the Open Access publication charges for this article was provided by the Canadian Institutes of Health Research (CIHR).
Conflict of interest statement. None declared.
REFERENCES
- 1.Baasiri RA, Glasser SR, Steffen DL, Wheeler DA. The breast cancer gene database: a collaborative information resource. Oncogene. 1999;18:7958–7965. doi: 10.1038/sj.onc.1203335. [DOI] [PubMed] [Google Scholar]
- 2.Hersh WR. Information Retrieval: A Health and Biomedical Perspective. 2nd. New York, NY: Springer; 2003. [Google Scholar]
- 3.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Church DM, DiCuccio M, Edgar R, Federhen S, Helmberg W, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2005;33(Database issue):D39–D45. doi: 10.1093/nar/gki062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gasteiger E, Jung E, Bairoch A. SWISS-PROT: connecting biological knowledge via a protein database. Curr. Issues Mol. Biol. 2001;3:47–55. [PubMed] [Google Scholar]
- 5.Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN. Human gene mutation database (HGMD®): 2003 update. Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 6.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34(Database issue):D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, et al. HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007;35(Database issue):D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanabe L, Scherf U, Smith LH, Lee JK, Hunter L, Weinstein JN. MedMiner: an Internet text-mining tool for biomedical information, with application to gene expression profiling. Biotechniques. 1999;27:1210–1217. doi: 10.2144/99276bc03. [DOI] [PubMed] [Google Scholar]
- 9.Hu Y, Hines LM, Weng H, Zuo D, Rivera M, Richardson A, LaBaer J. Analysis of genomic and proteomic data using advanced literature mining. J. Proteome Res. 2003;2:405–412. doi: 10.1021/pr0340227. [DOI] [PubMed] [Google Scholar]
- 10.Maier H, Dohr S, Grote K, O’Keeffe S, Werner T, Hrabe de Angelis M, Schneider R. LitMiner and WikiGene: identifying problem-related key players of gene regulation using publication abstracts. Nucleic Acids Res. 2005;33(Webserver issue):W779–W782. doi: 10.1093/nar/gki417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann R, Valencia A. Implementing the iHOP concept for navigation of biomedical literature. Bioinformatics. 2005;21(Suppl. 2):ii252–ii258. doi: 10.1093/bioinformatics/bti1142. [DOI] [PubMed] [Google Scholar]
- 12.Plake C, Schiemann T, Pankalla M, Hakenberg J, Leser U. Alibaba: PubMed as a graph. Bioinformatics. 2006;22:2444–2445. doi: 10.1093/bioinformatics/btl408. [DOI] [PubMed] [Google Scholar]
- 13.Rebholz-Schuhmann D, Kirsch H, Arregui M, Gaudan S, Riethoven M, Stoehr P. EBIMed—text crunching to gather facts for proteins from Medline. Bioinformatics. 2007;23:e237–e244. doi: 10.1093/bioinformatics/btl302. [DOI] [PubMed] [Google Scholar]
- 14.Hamosh A, Scott AF, Amberger J, Bocchini C, Valle D, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2002;30:52–55. doi: 10.1093/nar/30.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wain HM, Lush M, Ducluzeau F, Povey S. Genew: the human gene nomenclature database. Nucleic Acids Res. 2002;30:169–171. doi: 10.1093/nar/30.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mishra G, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P, Shivkumar K, Anuradha N, Reddy R, Raghavan TM, et al. Human Protein Reference Database—2006 update. Nucleic Acids Res. 2006;34(Database issue):D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Humphreys BL, Lindberg DA, Schoolman HM, Barnett GO. The unified medical language system: an information research collaboration. J. Am. Med. Inform. Assoc. 1998;5:1–13. doi: 10.1136/jamia.1998.0050001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becker KG, Barnes KC, Bright TJ, Wang SA. The Genetic Association Database. Nat. Genet. 2004;36:431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- 20.Riggins GJ, Strausberg RL. Genome and genetic resources from the Cancer Genome Anatomy Project. Hum. Mol. Genet. 2001;10:L663–L667. doi: 10.1093/hmg/10.7.663. [DOI] [PubMed] [Google Scholar]
- 21.Thorisson GA, Smith AV, Krishnan L, Stein LD. The International HapMap Project Web site. Genome Res. 2005;15:1592–1593. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hao Y, Zhu X, Huang M, Li M. Discovering patterns to extract protein–protein interactions from the literature: part II. Bioinformatics. 2005;21:3294–3300. doi: 10.1093/bioinformatics/bti493. [DOI] [PubMed] [Google Scholar]
- 23.Huang M, Zhu X, Hao Y, Payan DG, Qu K, Li M. Discovering patterns to extract protein–protein interactions from full texts. Bioinformatics. 2004;20:3604–3612. doi: 10.1093/bioinformatics/bth451. [DOI] [PubMed] [Google Scholar]