Abstract
The Superimposé webserver performs structural similarity searches with a preference towards 3D structure-based methods. Similarities can be detected between small molecules (e.g. drugs), parts of large structures (e.g. binding sites of proteins) and entire proteins. For this purpose, a number of algorithms were implemented and various databases are provided. Superimposé assists the user regarding the selection of a suitable combination of algorithm and database. After the computation on our server infrastructure, a visual assessment of the results is provided. The structure-based in silico screening for similar drug-like compounds enables the detection of scaffold-hoppers with putatively similar effects. The possibility to find similar binding sites can be of special interest in the functional analysis of proteins. The search for structurally similar proteins allows the detection of similar folds with different backbone topology. The Superimposé server is available at: http://bioinformatics.charite.de/superimpose.
INTRODUCTION
As the size of biomolecules differs by orders of magnitude, the ways to compare them and the metrics to measure what a good comparison actually is, often differ in the same respect. To cite Hugo Kubinyi: ‘Similarity lies in the eye of the beholder’ (1,2). Therefore, a classification of the alignment problem is required to determine the appropriate method for the detection of the similarity. The definition of similarity in molecular space always depends on the scientific question that is asked. This question heavily influences the design of the algorithm and the definition of the scoring function, which can be adjusted to fit the needs of each request. Unfortunately, comparison algorithms are computationally expensive since the problems are usually NP hard, which means that the retrieval of a result is at least extremely time consuming (3).
A number of algorithms as well as databases are free for non-commercial use, but in many cases there is no dedicated webserver that allows hassle-free use of an algorithm and a suitable database to answer a biological question. For small molecules, data sources such as PubChem (4) and Drugbank (5) provide facilities for similarity searching. In general, for small molecules their similarity is estimated on the basis of their chemical topology. One method is to translate the chemical topology into so called structural fingerprints. Structural fingerprints are bitvector representations of the small compound chemistry. To compare bitvectors of two molecules, metrical coefficients like the Tanimoto coefficent are applied. The Tanimoto coefficient gives values between 1.0 (very similar) and 0.0 (dissimilar) Another often used method is the representation of the molecule as string pattern (SMILES). A simple string search can be used to determine if a certain part of the molecule is present in another molecule or not. But a number of features of small molecules cannot be reflected adequately by 2D representations (6,7). Recent findings suggest that 3D similarity searches yield at least more varied results (8) than similarity comparisons via the usage of fingerprints or SMILES. Especially to find scaffold hoppers, 3D algorithms clearly show an advantage. For this reason, Superimposé is dedicated, but not limited to the usage of 3D algorithms.
There are a number of superposition servers, websites and projects in the field of protein similarity. Often they are merely a companion for a specific algorithm. For instance, the website of TM-align (9) allows to compare protein structures but not search depending on a database. Dedicated superposition servers for proteins include [10–13 and http://www.ncbi.nlm.nih.gov/Structure/VAST/]: 3dSS (10) has strengths by providing the ability to superimpose more than two proteins. Secondary structure matching (11) is a very fast method that even allows searches on a PDB scale level within minutes. However, due to the fact that algorithms in this field are often domain specific and have their own definitions of good matches, the possibility to choose among a set of algorithms would be beneficial. For a more comprehensive overview about macromolecular superposition, we recommend the reading of refs (14,15). For the problem of identifying a similar surface in or on macromolecules, there is no website that features such a service for the public yet. Such a service could help to elucidate similar functions of proteins based on shared binding sites or surface patches. Recent findings even suggest that similarities based on interaction patches of proteins can help to get hints about the docking modes between proteins (16).
For superposition tasks on Superimposé, we define a three class division of problem cases for molecular similarity searches that branch to different subtasks the user can solve with its help.
Similarity Class 1: Small molecule level.
Similarity Class 2: Macromolecule level based on substructures.
Similarity Class 3: Protein level.
Searches according to Class 1 and Class 3 aim at assigning as many atoms as possible between both structures. For small molecules (compounds), this often means that retrieved compounds are similar in mode of action and/or are affecting similar targets (17). Class 2 algorithms are assuming that the query structure is smaller than the macromolecule. A typical scenario for Class 2 algorithms is the identification of similar binding sites. Class 3 specially targets the comparison of entire proteins. The order of amino acids in the peptide chain is a valuable information in addition to the 3D coordinates. In most cases of pairs of homologous proteins, the corresponding amino acids appear in the same order. This is because the order of amino acids is preserved in evolution, unless it is disrupted by recombinatory events leading to circular permutation. However, the number of considered atoms is often reduced by different levels: C-alpha, backbone. Algorithms operating on the protein backbone or even on all-atom-level are often inefficient for protein comparisons (18). Established methods therefore often choose hierarchical approaches by dividing the protein into structural elements (19).
The preparation of databases, the installation of programs for structure comparison and the sorting and visual inspection of search results is often a complex task with currently available tools. Superimposé facilitates database searches by providing an uniform user interface for different programs, databases and scoring functions. Several databases for small molecules are joined to one comprehensive collection of 3D structures. Users of Superimposé do not have to solve technical problems and can concentrate on the biological problem.
ALGORITHMS
This alphabetically ordererd section gives practical descriptions of algorithms deployed by Superimposé. If not stated otherwise, Superimposé uses original binaries with default parameters for the algorithms.
GangstaLite
GANGSTA (19) is an algorithm for structural alignment of proteins and similarity search. GangstaLite is a specially drafted fast version for the Superimposé project. GANGSTA works in two stages: in the first stage, a mapping on the secondary structure elements is generated using a combinatorial approach that replaces the former genetic algorithm. In the second stage, individual residue pairs are assigned to create a maximum contact overlap.
GangstaLite is designed to detect similarities between proteins without using sequential informations. Therefore, cases of fold similartity without sequential similarity will be recognized. An example of circular permutation is presented in the case studies.
NeedleHaystack
NeedleHaystack (20) computes structural alignments of molecules as superpositions of sets of single atoms in the 3D space, where information on chemical connectivity and atom types is not necessarily considered. It is specially suited to scan a large molecule (target = haystack, up to 100 000 atoms) for the occurrence of a given molecular motif (model = needle) with a given tolerance level. It operates on the complete enumeration of superpositions of atom triples in both model and target, but radical pruning reduces the running time to seconds for a typical problem size, the search for a binding site in a protein surface. As NeedleHaystack is used for binding-site recognition, we are using the parameters -sk 0.25, -ad 1.35, -al 2, -to 60, -bd 1. Additionally, NeedleHaystack uses a weighting matrix that punishes each missed superposition on atom level with the score 2.
A typical application for this algorithm is the search for similar binding sites. This is illustrated in the Case studies section.
Point set match (PSM)
PSM (21) is a program that finds and aligns a small search pattern in a large search space, e.g. some sort of known substructure in a possibly large protein. PSM is an efficient implementation of a subgraph matching algorithm that uses certain domain-specific heuristics. The atoms represent the vertices of the distance graphs, their distances among each other represent the edges of the graph. The lengths of the edges of the distance graph over the search pattern are used to construct the distance graph over the search space, where only the edges that have similar lengths as the corresponding edges in the search pattern are maintained. With the help of a backtracking algorithm, PSM enumerates all possible matchings. Heuristics are used to order the vertices and edges during the search in such a way that the algorithm discards non-profitable partial matches early. The heuristics include, for instance, atom type, membership to a certain chemical group of the atoms and frequency of edge distance in the graphs. PSM not only finds the ideal alignment based on dRMS (distance root mean square), but also is able to compute the (locally) optimal alignment for average distance, maximum distance or any other distance metrics. PSM uses the derivative free minimization algorithms taken from ref. (22) to compute the rigid motion transformation, including a small scaling factor. Due to the fact that PSM is based on distance graphs, it can be easily extended to work with deformable search patterns where hinges and torsions are allowed. Furthermore, individual tolerances can be assigned to all edges and L-matches (i.e. mirrored matches) can be found. PSM is able to recognize similar surface patches/active sites.
Score1
For a partial superposition M (i.e. partial matching of atoms) between the two input molecules, the score of M is defined as follows:
| 1 |
where r is the proportion of superimposed non-hydrogen atoms of the smaller molecule and rmsd(M) is the square root of the least possible mean-squared distance between atom pairs matched in M under all possible rigid motions of the input molecules. Therefore, score∈(0.0,1.0] acts as a geometric similarity measure between two input molecules. If one molecule is identical to another molecule, then there is a superposition M such that score(M) = 1.0. Score1 calculates an optimal spatial superposition of two drug-sized molecules with respect to the above score function subject to an additional constraint: for every atom a matched in the superposition, there has to be an atom b bound to a such that b is matched, too. This restriction of the search space allows to use an optimal branch-and-bound algorithm as described in ref. (8) without any reduction of the input molecules. To speed up the algorithm, also lower bounds for possible solutions along different paths in the search tree are calculated. Promising paths can be searched first, leading to a more effective pruning. To establish the lower bounds, techniques from ref. (23) for calculating the optimum atom pairs given a fixed rigid motion of the input molecules are used. In accord with the authors, we are using the parameters ‘0.7 0.65 0.0’ that enables us to use Score1 in whole database screening applications.
Score1 is suitable for similarity screening in small molecule databases, illustrated in the case studies section.
sd_best_compare
The algorithm sd_best_compare is based on a normalization of the atomic sets according to their principal moments of inertia (24). This first normalization is of course independent of transformations of the coordinate system, and quite stable for small alterations of the atomic positions. It is also unique except for four possible rotations. Therefore, the degree of freedom is strongly reduced and the assignment of pairs of related atoms is straightforward for identical or very similar sets. In the first step, both atomic sets are roughly orientated according to their size proportions. After superimposing the centres of mass and alignment of the longest and smallest dimensions closest atoms are assigned as pairs. This assignment is improved by numerous refinement cycles. The algorithm was tailored for the search of similar atomic sets in a large database of patches (not necessarily bonded atoms) (25); the aim of the algorithm is not to compare very different molecules, but to find similar molecules with different connection schema. To do this as fast as possible, the database should be prepared to minimize the effort of parsing the data file (26). With the help of some adapted procedures the method can also be used to compare entire proteins.
The algorithm was implemented to compare conformational databases of low molecular weight structures that share similar scaffold (8).
TM-align
TM-align (9) uses a two-step process that is made up of an initial structural alignment based on a initial assignment of secondary structure element and dynamic programming. This step is followed by a heuristic optimization. The alignment as well as the heuristic optimization is based on TM-score. TM-score is a variation of the Levitt–Gerstein weight factor that punished larger distances relatively stronger than smaller distances and allows more sensitivity concerning the global topology. The value of TM-score lies in (0,1]. In general, a comparison of score <0.2 indicates that there is no similarity between two structures; generally, a TM-score >0.5 indicates that structures share the same fold, but the drop-off of the score indicating the twilight-zone of similarity has to be considered individually. TM-align is an algorithm for protein structure alignment.
CE (combinatorial extension)
The algorithm CE (27) involves a combinatorial extension of an alignment path defined by aligned fragment pairs (AFP), which represent possible alignment paths. Combinations of AFPs are selectively extended or discarded to yield an optimal alignment path. They are based on local geometry, rather than global features such as orientation of secondary structures and overall topology. The algorithm is fast and accurate in elucidating structural alignments and fast enough for database scanning and detailed analyses of protein families. CE builds an alignment between two protein structures.
DATABASES
This section provides information about the databases in alphabetical order. Databases are updated on a monthly basis.
Astral 40
The Astral Compendium (28) provides several databases and tools derived partly from the SCOP (29) database and based on PDB coordinate files. SCOP itself provides schemas of all proteins available in the PDB according to their evolutionary and structural relationships. Additionally, a grouping of proteins into species and a classification into families and superfamilies, folds and classes is provided. ASTRAL 40 provides this information filtered with 40% sequence identity in a PDB style format that is deployed onto the Superimposé webserver. Astral provides 9500+chains/domains and aims to represent the whole structural space of proteins. We are providing a link to the PDBSum (30) that enables the user to examine the found proteins in great detail with the original paper.
Ligand Depot
The Ligand Depot (31) is a data warehouse that integrates databases, services, tools and methods related to small molecules bound to macromolecules. It provides chemical and structural information about small molecules in entries of the Protein Data Bank. Currently, it contains information about 80 000+ structures. All small structures of the Ligand Depot are deployed on the Superimpose server and allow to search for the occurrence of small molecules or analogues in the PDB.
Open NCI database
The release of the Open NCI Database (32) includes 210 000+ compounds with 25 conformers on average. The Open NCI database contains compounds that show a significant activity as therapeutic agent against diseases like AIDS and cancer. A molecule that is highly similar to a compound in the Open NCI might have similar medical activities. For further investigation, we are providing a link to the Enhanced NCI Database Browser (33).
PDB (Culled)
The PDB (34) is an archive of experimentally determined, biological macromolecule 3D structures and contains 48 500+ structures of proteins. Because of the nature of the PDB as all purpose repository for macromolecules it often contains duplicate structures and structures of a resolution that are hardly suitable for searching. Another problem is the sheer size of the PDB, what makes it impossible for many algorithms to perform comparisons between proteins (Class 3) and on substructures of proteins (Class 2). For both the reasons, we are using a representative subset of the PDB. The subset is calculated using the PISCES Server (35). The used cut-off thresholds are: sequence identity cut-off: 20%; resolution cut-off: 1.8A and 2.2A; R-factor cut-off: 0.25. A link to the PDBSum is provided.
PDB surfaces (Culled)
For the elucidation of similar parts on the surfaces of macromolecules, it is suitable to limit the search space to the water accessible surface. None of the presented algorithms does this on its own, so a pre-computing step is applied for the PDB (Culled) Database described above. We are using an algorithm calc-surface (36) to generate macromolecules with the water accessible surface alone. A link to the PDBSum is provided.
Superdrug
The Superdrug (37) database contains 2500+ 3D structures of active ingredients of essential marketed drugs. To account for structural flexibility, they are represented on average by about 40 structural conformers per drug generated by the program Catalyst (Accelrys Inc. http://www.accelrys.com). Superdrug provides a link to the Superdrug website that enables the user to investigate results in more detail like the ATC code (WHO classification of medical compounds according to their therapeutic application and chemical scaffold).
WEBSERVER DESCRIPTION
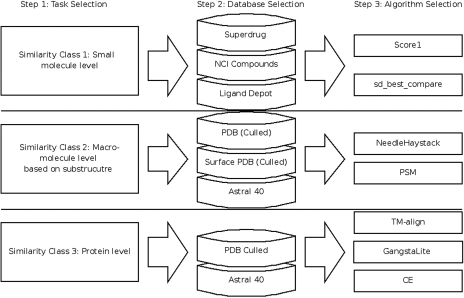
For Superimposé, we decided to provide a wizard style approach that guides the user through the different possibilities we offer (Figure 1). We are using a fixed set of parameters for all algorithms that allow a generalized execution of task. A typical search workflow begins with the selection of a task the user wants to execute. This task maps to the three classes described in the Introduction section. In the next step, the user can upload a file to act as model (or patch in Class 2) for the search. Supported file formats are sdf, mol and pdb. Conversions between different file formats are handled via OpenBabel (38). Subsequently, the user gets a selection of suitable databases and algorithms for that task.
Figure 1.
Suitable combinations of databases with algorithms depending on the class of the scientific problem.
Computations can take longer times (24 h) in case where there are several users employing the web service. Therefore, the user provides an email address, where a report about finished jobs is directed to. This email contains a hyperlink to a webpage on the Superimposé server that presents all results for the search with possibilities to visually assess the results. We are providing a specially designed visualization via Jmol as a Java Applet. This allows the user to execute custom scripts in the Jmol language for extensive visualization. The second visualization possibility especially tailored for proteins is STRAP (39), which is implemented via Java Webstart and behaves like a native application and not like a webpage as Jmol does. For both programs, the sole requirement is a Java JRE (http://java.com).
CASE STUDIES
The following case studies are organized per problem class and show typical problems where Superimposé can be applied. All molecules and proteins that are discussed within the case studies are available for download on the Superimposé web page (documentation).
Small structure similarity (Class 1)
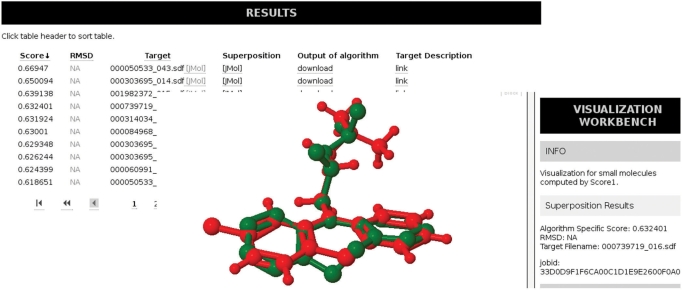
Similar compounds are more or less likely to share properties such as ligand specificity and binding strength. Thus, screening for similar compounds in databases is a standard technique to generate new hypotheses for molecules (shared activity). Therefore, Superimposé allows the user to search for similarities against a variety of compound databases. In this case, we want to highlight the ability of Superimposé to successfully retrieve similar compounds to Chlorpromazine (ATC: N05AA01) on the database Superdrug with the algorithm Score1. We define similarity as the ability to find compounds in a related ATC group. The results for the first 10 entries show that Superimposé is able to find compounds that are apart from two compounds Methdilazine (ATC: R06AD04) and Pimethixene (ATC: R06AX23), all coming from the desired ATC-code N (Nervous System). For the two compounds from ATC group R (Respiratory System), this could point to unwanted side-effects of Chlorpromazine. The fingerprint-based search on the website of Superdrug fails in retrieving the compounds Trimipramine (ATC: N06AA06) and Cyamemazine (ATC: N05AA06).
Compared with the results of the Superdrug website Superimposé is additionally able to successfully retrieve compounds Trimipramine (ATC-code N06AA06) and Cyamemazine (ATC-code N05AA06), which are left out by the fingerprint search. The reason is that structural superposition is able to to superimpose scaffold hoppers, in this case a six- and seven-membered ring structure (Figure 2), which are dissimilar in the SuperDrug fingerprint search.
Figure 2.
Query compound Chlorpromazine(red) and search hit Trimipramine (green).
Substructure search (Class 2)
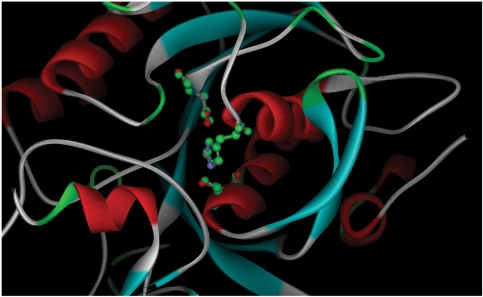
Here, we want to show the ability of the NeedleHaystack algorithm together with the CulledPDB to identify related proteins based on a patch from the catalytic site. For the case study, we are using a patch from the active site of protein Hydrolase (PDB-code: 1PEK). This patch is successfully identified on a Subtilisin complex (PDB-code: 2SIC) with related activity. NeedleHaystack retrieves perfect matches, e.g. in the active site of 2SIC (Figure 3). entries
Figure 3.
Superposition of the active site derived from protein Hydrolase (PDB-code: 1PEK/green atoms) that is successfully identified in protein Subtilisin (PDB-code: 2SIC/cpk coloured ball and sticks in the middle).
Protein similarity (Class 3)
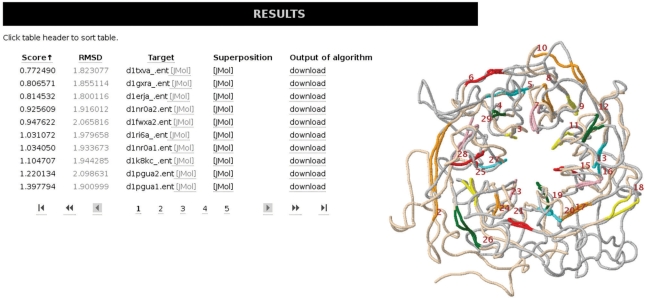
For the problem of protein similarity/protein alignment, a main case where sequence-based methods often fail is for proteins that are similar in terms of overall structure (fold) but not on sequence level. One example where especially the GangstaLite algorithm can find meaningful alignments is a Integrin alpha-V (PDB-code: 1M1X). In combination with the Astral database, GangstaLite successfully retrieves a WD40 domain of the Transcriptional Repressor TUP1 (PDB-code: 1ERJ) as one of the best scoring alignments (Figure 4). GangstaLite successfully aligns the proteins with half of the secondary elements not in sequence direction.
Figure 4.
Results (left) and non-sequential structural alignment generated by GangstaLite (right).
CONCLUSIONS
Superimposé is created to deal with structural superpositions of molecules in a widespread sense. The combination of databases and algorithms of different fields provides amongst others the possibility to identify similar proteins, similar medical active compounds and also binding-sites via similarities in substructure search. The server will be useful for bioinformaticians who have specialized on structures, macromolecular biologists and the systems biology community by providing possibilities to identify similar patches (binding sites/surface patches) in known proteins. By reducing the complexity of installing algorithms, databases and finding suitable parameter sets Superimposé allows researchers to instantly deal with the task without the administrative problems around it.
A major upgrade of Superimposé is planned for the end of 2008, where we will deploy more algorithms and databases on the server. We are aiming to incorporate many of the feature requests of the community and appropriately extend the server.
ACKNOWLEDGEMENTS
We thank Yang Zhang for the permission to use TM-align on the Superimposé server and also Igor Filippov, Marc C. Nicklaus and Wolf-Dietrich Ihlenfeldt from the NCI for providing data of the Open NCI Database with conformers. This effort is supported by DFG (Deutsche Forschungsgemeinschaft) SFB-449, Deutsche Krebshilfe and the DFG International Research Training Group (IRTG) on ‘Genomics and Systems Biology of Molecular Networks’ (GRK1360). We also thank the Charité CBF computing centre for their support. Without the use of free and/or open source software cited or running quietly on the backend of the server this effort would not have been possible. In this regard, we especially want to thank the developers of Jmol (http://jmol.org), CDK (http://cdk.sf.net) and OpenBabel (http://openbabel.sf.net). Funding to pay the Open Access Publication charges for this article was provided by Deutsche Forschungsgemeinschaft (SFB-449).
Conflict of interest statement. None declared.
REFERENCES
- 1.Kubinyi H. [Molecular similarity. 2. The structural basis of drug design] Pharm. Unserer Zeit. 1998;27:158–172. doi: 10.1002/pauz.19980270406. [DOI] [PubMed] [Google Scholar]
- 2.Kubinyi H. [Molecular similarity. 1. Chemical structure and biological action] Pharm. Unserer Zeit. 1998;27:92–106. doi: 10.1002/pauz.19980270305. [DOI] [PubMed] [Google Scholar]
- 3.Lathrop RH. The protein threading problem with sequence amino acid interaction preferences is np-complete. Protein Eng. 1994;7:1059–1068. doi: 10.1093/protein/7.9.1059. [DOI] [PubMed] [Google Scholar]
- 4.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2008;36 doi: 10.1093/nar/gkm1000. (Database Issue), D13–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. Drugbank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36 doi: 10.1093/nar/gkm958. (Database Issue), D901–D906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whittle M, Willett P, Klaffke W, vanNoort P. Evaluation of similarity measures for searching the dictionary of natural products database. J. Chem. Inf. Comput. Sci. 2003;43:449–457. doi: 10.1021/ci025591m. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, Reynolds CH. Performance of similarity measures in 2d fragment-based similarity searching: comparison of structural descriptors and similarity coefficients. J. Chem. Inf. Comput. Sci. 2002;42:1407–1414. doi: 10.1021/ci025531g. [DOI] [PubMed] [Google Scholar]
- 8.Thimm M, Goede A, Hougardy S, Preissner R. Comparison of 2d similarity and 3d superposition. Application to searching a conformational drug database. J. Chem. Inf. Comput. Sci. 2004;44:1816–1822. doi: 10.1021/ci049920h. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Skolnick J. Tm-align: a protein structure alignment algorithm based on the Tm-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sumathi K, Ananthalakshmi P, Roshan MNAM, Sekar K. 3dss: 3d structural superposition. Nucleic Acids Res. 2006;34 doi: 10.1093/nar/gkl036. (Web Server Issue), W128–W132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krissinel E, Henrick K. Secondary-structure matching (ssm), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallgr. 2004;60 doi: 10.1107/S0907444904026460. (Pt 12 Pt 1), 2256–2268. [DOI] [PubMed] [Google Scholar]
- 12.Maiti R, Domselaar GHV, Zhang H, Wishart DS. Superpose: a simple server for sophisticated structural superposition. Nucleic Acids Res. 2004;32 doi: 10.1093/nar/gkh477. (Web Server Issue), W590–W594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leslin CM, Abyzov A, Ilyin VA. Topofit-db, a database of protein structural alignments based on the topofit method. Nucleic Acids Res. 2007;35 doi: 10.1093/nar/gkl809. (Database Issue), D317–D321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Novotny M, Madsen D, Kleywegt GJ. Evaluation of protein fold comparison servers. Proteins. 2004;54:260–270. doi: 10.1002/prot.10553. [DOI] [PubMed] [Google Scholar]
- 15.Kolodny R, Koehl P, Levitt M. Comprehensive evaluation of protein structure alignment methods: scoring by geometric measures. J. Mol. Biol. 2005;346:1173–1188. doi: 10.1016/j.jmb.2004.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Günther S, May P, Hoppe A, Frömmel C, Preissner R. Docking without docking: Isearch–prediction of interactions using known interfaces. Proteins. 2007;69:839–844. doi: 10.1002/prot.21746. [DOI] [PubMed] [Google Scholar]
- 17.Barbosa F, Horvath D. Molecular similarity and property similarity. Curr. Top. Med. Chem. 2004;4:589–600. doi: 10.2174/1568026043451186. [DOI] [PubMed] [Google Scholar]
- 18.Shakhnovich E. Protein folding thermodynamics and dynamics: where physics, chemistry, and biology meet. Chem. Rev. 2006;106:1559–1588. doi: 10.1021/cr040425u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolbeck B, May P, Schmidt-Goenner T, Steinke T, Knapp E-W. Connectivity independent protein-structure alignment: a hierarchical approach. BMC Bioinform. 2006;7:510. doi: 10.1186/1471-2105-7-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoppe A, Froemmel C. Needlehaystack: a program for the rapid recognition of local structures in large sets of atomic coordinates. J. Appl. Cryst. 2003;36:1090–1097. [Google Scholar]
- 21.Formella A. Approximate point set match for partial protein structure alignment. In: Couto FM, Silva JS, Fernandes P, editors. Proceedings of Bioinformatics: Knowledge Discovery in Biology (BKDB2005) Facultade Ciencias Lisboa da Universidade de Lisboa; 2005. pp. 53–57. [Google Scholar]
- 22.García-Palomares U, Rodríguez J. New sequential and parallel derivative–free algorithms for unconstraint optimization. SIAM J. Optim. 2002;13:79–96. [Google Scholar]
- 23.Kirchner S. An fptas for computing the similarity of three-dimensional point sets. Int. J. Comput. Geom. Appl. 2007;17:161–174. [Google Scholar]
- 24.Preissner R, Goede A, Frömmel C. Homonyms and synonyms in the dictionary of interfaces in proteins (dip) Bioinformatics. 1999;15:832–836. doi: 10.1093/bioinformatics/15.10.832. [DOI] [PubMed] [Google Scholar]
- 25.Frmmel C, Gille C, Goede A, Grpl C, Hougardy S, Nierhoff T, Preissner R, Thimm M. Accelerating screening of 3d protein data with a graph theoretical approach. Bioinformatics. 2003;19:2442–2447. doi: 10.1093/bioinformatics/btg343. [DOI] [PubMed] [Google Scholar]
- 26.Preissner R, Goede A, Rother K, Osterkamp F, Koert U, Froemmel C. Matching organic libraries with protein-substructures. J. Comput. Aided. Mol. Des. 2001;15:811–817. doi: 10.1023/a:1013158818807. [DOI] [PubMed] [Google Scholar]
- 27.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (ce) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 28.Chandonia J-M, Hon G, Walker NS, Conte LL, Koehl P, Levitt M, Brenner SE. The astral compendium in 2004. Nucleic Acids Res. 2004;32 doi: 10.1093/nar/gkh034. (Database Issue), D189–D192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conte LL, Brenner SE, Hubbard TJP, Chothia C, Murzin AG. Scop database in 2002: refinements accommodate structural genomics. Nucleic Acids Res. 2002;30:264–267. doi: 10.1093/nar/30.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laskowski RA. Enhancing the functional annotation of pdb structures in pdbsum using key figures extracted from the literature. Bioinformatics. 2007;23:1824–1827. doi: 10.1093/bioinformatics/btm085. [DOI] [PubMed] [Google Scholar]
- 31.Feng Z, Chen L, Maddula H, Akcan O, Oughtred R, Berman HM, Westbrook J. Ligand depot: a data warehouse for ligands bound to macromolecules. Bioinformatics. 2004;20:2153–2155. doi: 10.1093/bioinformatics/bth214. [DOI] [PubMed] [Google Scholar]
- 32.Voigt JH, Bienfait B, Wang S, Nicklaus MC. Comparison of the nci open database with seven large chemical structural databases. J. Chem. Inf. Comput. Sci. 2001;41:702–712. doi: 10.1021/ci000150t. [DOI] [PubMed] [Google Scholar]
- 33.Ihlenfeldt W-D, Voigt JH, Bienfait B, Oellien F, Nicklaus MC. Enhanced cactvs browser of the open nci database. J. Chem. Inf. Comput. Sci. 2002;42:46–57. doi: 10.1021/ci010056s. [DOI] [PubMed] [Google Scholar]
- 34.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang G, Dunbrack RL. Pisces: recent improvements to a pdb sequence culling server. Nucleic Acids Res. 2005;33 doi: 10.1093/nar/gki402. (Web Server Issue), W94–W98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai J, Taylor R, Chothia C, Gerstein M. The packing density in proteins: standard radii and volumes. J. Mol. Biol. 1999;290:253–266. doi: 10.1006/jmbi.1999.2829. [DOI] [PubMed] [Google Scholar]
- 37.Goede A, Jaeger IS, Preissner R. Superficial–surface mapping of proteins via structure-based peptide library design. BMC Bioinform. 2005;6:223. doi: 10.1186/1471-2105-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guha R, Howard MT, Hutchison GR, Murray-Rust P, Rzepa H, Steinbeck C, Wegner JK, Willighagen EL. The blue obelisk–interoperability in chemical informatics. J. Chem. Inf. Model. 2006;46:991–998. doi: 10.1021/ci050400b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gille C, Frömmel C. Strap: editor for structural alignments of proteins. Bioinformatics. 2001;17:377–378. doi: 10.1093/bioinformatics/17.4.377. [DOI] [PubMed] [Google Scholar]