Abstract
Many small RNA (sRNA) genes in bacteria act as posttranscriptional regulators of target messenger RNAs. Here, we present TargetRNA, a web tool for predicting mRNA targets of sRNA action in bacteria. TargetRNA takes as input a genomic sequence that may correspond to an sRNA gene. TargetRNA then uses a dynamic programming algorithm to search each annotated message in a specified genome for mRNAs that evince basepair-binding potential to the input sRNA sequence. Based on the calculated basepair-binding potential of each message with the given sRNA regulator, TargetRNA outputs a ranked list of candidate mRNA targets along with the predicted basepairing interaction of each target to the sRNA. The predictive performance of TargetRNA has been validated experimentally in several bacterial organisms. TargetRNA is freely available at http://snowwhite.wellesley.edu/targetRNA.
INTRODUCTION
In bacteria, the number of characterized small, noncoding RNAs (sRNAs) that have intrinsic functions as regulators has steadily increased in recent years. Many sRNAs act by posttranscriptionally regulating mRNAs via basepairing interactions (1). In Escherichia coli, all of the sRNAs that act by basepairing affect either the stability or translation of the mRNA target. In most cases, the mRNA targets of sRNA regulation are trans-encoded, and the sRNA:mRNA basepairing interactions are interrupted by gaps in the pairing. Further, most sRNAs in this class bind to the RNA chaperone Hfq, which has been shown to facilitate the interaction between many sRNAs and their targets (2).
Here, we describe a program, TargetRNA, which can effectively predict mRNA targets of basepairing sRNAs. While a number of approaches have been described for identifying targets of microRNA genes in eukaryotes (3–7), there have been relatively few computational tools developed for characterizing targets of sRNA regulators in bacteria. Mandin et al. (8) describe one such tool, which has been applied successfully in the genome of Listeria monocytogenes, though the program is not available as a webserver. Vogel and Wagner (9) offer a comprehensive review of approaches, both computational and experimental, for identifying targets of bacterial sRNAs. The program we describe here, TargetRNA, is accessible via a webserver that has been operating and publicly available since 2005, processing approximately 1000 sequence submissions per month. The webserver consists of a 16 CPU parallel computing cluster, and TargetRNAs underlying search algorithm has been designed for parallel computation, in order to increase performance and to process efficiently a large number of sequence submissions. The predictive performance of TargetRNA has been validated experimentally in E. coli using both northern blot and microarray experiments (10). Additionally, predictions from TargetRNA have been validated in other organisms, such as in Vibrio cholerae (11) and in Neisseria meningitidis (12), or are consistent with experimentally validated interactions, such as in Salmonella (13).
TargetRNA WEBSERVER
TargetRNA takes, as input, a genomic sequence that may correspond to a sRNA gene. TargetRNA then searches each annotated message in the user-specified genome for mRNAs that evince statistically significant basepair-binding potential to the input sRNA sequence. Basepair-binding potential of a sRNA with each mRNA is determined using one of two user-selected hybridization scoring methods. A detailed description of the hybridization scoring methods has been described previously (10). In summary, the individual basepair model of hybridization scoring is analogous to the Smith–Waterman dynamic program (14), except that instead of assessing homology potential, basepairing potential is assessed. The individual basepair model of hybridization scoring is the default scoring model for TargetRNA. Alternatively, a user can select a stacked basepair model for hybridization scoring. The stacked basepair model of hybridization scoring is based on stacking and destabilizing energies of interacting sequences. The stacked basepair model calculates the minimum free energy of hybridization for two RNA sequences, without allowing intramolecular basepairings. This model for hybridization scoring closely follows that developed and used for the RNAhybrid algorithm (7). The stacked basepair model is computationally more intensive than the individual basepair model and, as a result, increases the time required for calculating basepair binding potential by a factor of approximately five for most sequence inputs.
Once TargetRNA determines hybridization scores for the sRNA sequence with each message in a genome, the statistical significance of each potential sRNA:mRNA interaction is assessed. Determination of statistical significance is similar to that described for the RNAhybrid algorithm (7). Ten thousand random RNA sequences are generated such that the nucleotides in the random sequences are drawn from the distribution of nucleotides in the actual mRNA search space. The hybridization score is computed for each of these random sequences and the sRNA sequence. The resulting distribution of ten thousand hybridization scores is used to estimate a P-value for an sRNA:mRNA hybridization score by determining the probability of observing a score, by chance, equal to or less than the given sRNA:mRNA hybridization score.
After computing the statistical significance of basepair binding between a sRNA sequence and each message in a genome, TargetRNA outputs a ranked list of mRNAs in a genome whose basepair-binding potential with the sRNA sequence meets a significance threshold (the default is a P-value ≤ 0.01). Messages with statistically significant basepair-binding potential are considered candidate targets of sRNA regulation. Figure 1 illustrates example output from TargetRNA when searching for mRNA targets of the sRNA gene Spot42 in E. coli (15). As shown in Figure 1, TargetRNA outputs, for each candidate mRNA target identified, an annotation of the mRNA, a visual representation of its predicted basepair binding with the sRNA, and a P-value corresponding to the significance of the hybridization score of the predicted sRNA:mRNA interaction. To facilitate further investigation of candidate message targets, each identified target is linked to the Entrez Gene database (16) from the National Center for Biotechnology Information. The time required for TargetRNA to process an input sequence and generate results depends on the length of the sequence and the parameter selection but typically takes only a few seconds. Output from TargetRNA is available via both a formatted web page and a text file.
Figure 1.
The figure illustrates example output from using the TargetRNA webserver with default parameter settings to search for message targets of the sRNA Spot42 in Escherichia coli. In the middle of the figure, the six message targets predicted by TargetRNA are summarized. In the bottom of the figure, the predicted interaction between Spot42 and one of the six predicted targets, galK, is illustrated. The predicted interaction between Spot42 and galK consists of 41 nucleotides (from nucleotide 21 to 61) in the 109 nucleotide sRNA Spot42, and 39 nucleotides (from 19 nucleotides upstream of the galK start codon to 20 nucleotides downstream in the galK coding sequence) in the galK message.
The TargetRNA webserver provides a number of advanced search options that allow users more flexible control over the target search space beyond that provided by the default parameter settings. TargetRNA provides the option of automatically identifying and removing the region of the sRNA sequence corresponding to the terminator stem-loop since, in many cases, the terminator stem-loop does not participate in the sRNA:mRNA interaction. Users have the option of focusing their search around the 5′ UTR or 3′ UTR of messages, specifying the number of nucleotides to include upstream and downstream of the messages’ start codon or stop codon. Specifying regions around the 5′ UTR to be searched may be advantageous since many documented target interactions occur in particular regions, such as around the ribosome-binding sites, of messages. A seed, which corresponds to a minimum required length for at least one stretch of consecutive basepaired nucleotides in the sRNA:mRNA interaction, can also be specified. The seed is meant to reflect, biologically, the initial interaction between sRNA and mRNA, which has been shown in some cases to be a stretch of unpaired nucleotides in a loop of the sRNA that first basepairs with the target message. While TargetRNA searches each annotated message in a genome by default, users have the option of searching an individual message in order to explore more carefully a particular interaction predicted by TargetRNA. When searching for targets of a sRNA in a given organism, TargetRNA also offers the option of calculating the hybridization scores of orthologous targets with orthologous sRNAs in other organisms. Since many sRNA genes are conserved across related species, the program can thus suggest whether it is likely that the targets and hybridization interactions are conserved.
The various user-adjustable program parameters have the benefit of allowing a user to explore the tradeoff between sensitivity and specificity when assessing results of the TargetRNA program. For instance, removing the sRNA terminator sequence, focusing searches for targets to regions around translation start sites of messages, and setting a seed threshold above about five nucleotides can eliminate many false positive predictions. However, too much stringency with these parameters may result in a lack of identification of true targets. For example, some sRNA:mRNA interactions include the terminator sequence of the sRNA, such as the OxyS:fhlA interaction (17,18) in E. coli, and some sRNAs interact with messages somewhat distant from the message's start of translation, such as DsrA (19) and RprA (20) when interacting with rpoS in E. coli. Thus, the default parameter settings provide only a starting point for message target investigation. Default parameter settings were determined by optimizing performance of TargetRNA on sRNA:mRNA interactions in E. coli reported prior to 2005. Considering the recent growth in the number of sRNA:mRNA interactions reported in the literature across a range of bacterial species, we have revisited the issue of whether different default parameter settings would improve the performance of TargetRNA. When accounting for a broad set of sRNA:mRNA interactions in different species, we did not find a set of parameters that led to significant improvement, over the current default parameter settings, in successfully identifying targets of sRNA regulation.
RNATarget
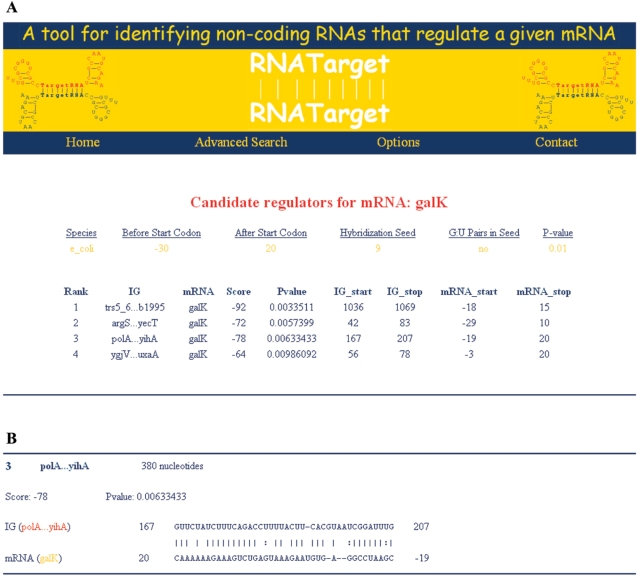
A recent addition to the TargetRNA webserver is a companion program to TargetRNA called RNATarget, which provides ‘reverse’ searching capabilities. Whereas TargetRNA takes as input the sequence of a candidate sRNA region and searches the genome for possible message targets, RNATarget takes as input the sequence of a candidate message target of an unknown sRNA regulator and searches the genome for regions containing possible sRNA regulators of the target sequence. RNATarget searches all intergenic sequences greater than 50 nucleotides in length in the genome for regions that evince significant basepair-binding potential to the input candidate target sequence. RNATarget outputs a ranked list of intergenic regions in the genome whose basepair-binding potential with the target sequence meets a significance threshold (the default is a P-value ≤ 0.01). Intergenic regions with statistically significant basepair-binding potential are considered as candidate regions corresponding to a sRNA regulator. Figure 2 illustrates example output from RNATarget when searching intergenic regions for candidate sRNA regulators of the mRNA target galK in E. coli. It has been reported previously that the sRNA Spot42, which resides in the intergenic region of the genome between genes polA and yihA, interacts with and regulates galK (21). As shown in Figure 2, RNATarget outputs, for each intergenic region identified, the flanking protein-coding genes, a visual representation of the predicted basepair binding between the two genomic sequences, and a P-value corresponding to the significance of the hybridization score of the predicted mRNA:sRNA interaction. Output is available via both a formatted web page and a text file. It is worth noting that RNATarget searches only for genomic regions demonstrating basepair-binding potential to the target sequence, it does not attempt to identify other properties of genomic sequences suggestive of sRNA genes, such as transcription initiation or termination sequences. Several other computational programs are available for predicting sRNA genes in a genome based on various sources of evidence, including transcription signals and comparative genomics information (22).
Figure 2.
The figure illustrates example output from using the RNATarget program with default parameter settings to search for intergenic regions in E. coli that evince basepair binding potential with a region of the galK message around its ribosome binding site. (A) A summary of four intergenic regions predicted by RNATarget is shown. (B) Details of the predicted interaction between galK and one of the intergenic regions, between genes polA and yihA, is shown. The sRNA Spot42, which resides in this intergenic region, is known to interact with and regulate galK (21). The predicted interaction between the intergenic region and galK consists of 41 nucleotides (from nucleotide 167 to 207) in the 380 nucleotide polA … yihA intergenic region, and 39 nucleotides (from 19 nucleotides upstream of the galK start codon to 20 nucleotides downstream in the galK coding sequence) in the galK message.
DISCUSSION AND CONCLUSIONS
TargetRNA is a freely available webserver that predicts message targets of sRNA action in bacteria. Predictions from TargetRNA have been validated experimentally in several organisms. Assessment of TargetRNA's performance suggests that the predictive performance of TargetRNA varies between sRNAs. For some sRNA regulators where one or more targets have been reported in the literature, TargetRNA successfully identifies the majority of targets, whereas for other sRNA regulators with one or more targets, TargetRNA identifies few, if any, targets (10). TargetRNA operates under the assumption that the basepair-binding potential of two genomic sequences, corresponding to an sRNA and a mRNA target, can serve as a predictor, albeit imperfect, for the interaction of the two RNAs. Conserved basepair-binding potential of two genomic sequences across different genomes can provide further evidence for sRNA:mRNA interaction. TargetRNA provides a feature to facilitate investigation of orthologous sRNA:mRNA interactions. However, other factors that may contribute to sRNA:mRNA interactions, such as RNA secondary structure or the role of Hfq, are not modeled by the program. As more examples of sRNA:mRNA interactions are reported across a range of bacteria, we will gain a better understanding of the various RNA properties and components within the cell that contribute to the interactions, and computational methods that aid in investigation of these interactions can be evolved appropriately to incorporate the new insights.
ACKNOWLEDGEMENTS
We thank Gisela Storz and Susan Gottesman for motivating the development of this work, for experimental validation of many computational predictions and for thoughtful discussions regarding its evolution. This work is supported in part by the National Institute of General Medical Sciences under grant GM078080. Funding to pay the Open Access publication charges for this article was provided by a Brachman Hoffman grant at Wellesley College.
Conflict of interest statement. None declared.
REFERENCES
- 1.Storz G, Gottesman S. In: The RNA World. 3rd. Gesteland RF, Cech TR, Atkins JF, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 567–594. [Google Scholar]
- 2.Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 3.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA – target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grimson A, Farh K.KH, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajewsky N. microRNA target predictions in animals. Nat. Genet. 2006;38:S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 6.Krek A, Grun D, Poy MN, Wolf R, Rosenburg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 7.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mandin P, Repoila F, Vergassola M, Geissmann T, Cossart P. Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res. 2007;35:962–974. doi: 10.1093/nar/gkl1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogel J, Wagner E.GH. Target identification of small noncoding RNAs in bacteria. Curr. Opin. Microbiol. 2007;10:262–270. doi: 10.1016/j.mib.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Tjaden B, Goodwin SS, Opdyke JA, Guillier M, Fu DX, Gottesman S, Storz G. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res. 2006;34:2791–2802. doi: 10.1093/nar/gkl356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis BM, Waldor MK. Rnase E-dependent processing stabilizes MicX, a Vibrio cholerae sRNA. Mol. Microbiol. 2007;65:373–385. doi: 10.1111/j.1365-2958.2007.05796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mellin JR, Goswami S, Grogan S, Tjaden B, Genco CA. A novel Fur- and iron-regulated small RNA, NerrF, is required for indirect Fur-mediated regulation of the sdhA and sdhC genes in Neisseria meningitidis. J. Bacteriol. 2007;189:3686–3694. doi: 10.1128/JB.01890-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007;21:2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 15.Sahagan B, Dahlberg JE. A small, unstable RNA molecule of Escherichia coli: Spot 42 RNA. J. Mol. Biol. 1979;131:593–605. doi: 10.1016/0022-2836(79)90009-3. [DOI] [PubMed] [Google Scholar]
- 16.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altuvia S, Zhang A, Argaman L, Tiwari A, Storz G. The Escherichia coli oxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J. 1998;17:6069–6075. doi: 10.1093/emboj/17.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Argaman L, Altuvia S. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Mol. Biol. 2000;300:1101–1112. doi: 10.1006/jmbi.2000.3942. [DOI] [PubMed] [Google Scholar]
- 19.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl Acad. Sci. USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majdalani N, Hernandez D, Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 2002;46:813–826. doi: 10.1046/j.1365-2958.2002.03203.x. [DOI] [PubMed] [Google Scholar]
- 21.Moller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 2002;16:1696–1706. doi: 10.1101/gad.231702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livny J, Waldor MK. Identification of small RNAs in diverse bacterial species. Curr. Opin. Microbiol. 2007;10:96–101. doi: 10.1016/j.mib.2007.03.005. [DOI] [PubMed] [Google Scholar]