Abstract
PROTEUS2 is a web server designed to support comprehensive protein structure prediction and structure-based annotation. PROTEUS2 accepts either single sequences (for directed studies) or multiple sequences (for whole proteome annotation) and predicts the secondary and, if possible, tertiary structure of the query protein(s). Unlike most other tools or servers, PROTEUS2 bundles signal peptide identification, transmembrane helix prediction, transmembrane β-strand prediction, secondary structure prediction (for soluble proteins) and homology modeling (i.e. 3D structure generation) into a single prediction pipeline. Using a combination of progressive multi-sequence alignment, structure-based mapping, hidden Markov models, multi-component neural nets and up-to-date databases of known secondary structure assignments, PROTEUS is able to achieve among the highest reported levels of predictive accuracy for signal peptides (Q2 = 94%), membrane spanning helices (Q2 = 87%) and secondary structure (Q3 score of 81.3%). PROTEUS2's homology modeling services also provide high quality 3D models that compare favorably with those generated by SWISS-MODEL and 3D JigSaw (within 0.2 Å RMSD). The average PROTEUS2 prediction takes ∼3 min per query sequence. The PROTEUS2 server along with source code for many of its modules is accessible a http://wishart.biology.ualberta.ca/proteus2.
INTRODUCTION
Ten years ago, the sequencing of whole genomes was a formidable, multi-year challenge. Now, thanks to advances in DNA sequencing technology, it is possible to sequence an entire bacterial genome in as little as a week (1). It is clear that our capacity to sequence organisms far outpaces our capacity to manually annotate their genomes (2). As a result, there is a growing interest in developing software to facilitate automated or semi-automated genome annotation (3). At the same time, there is an increasing desire to develop automated methods that can generate comprehensive annotations—annotations that provide detailed information about each protein's function, location, interacting partners, substrates, pathways and structure. Our laboratory has a long-standing interest in developing comprehensive, automated genome/proteome annotation tools (3–5). We also believe that high-quality structure prediction and modeling can play an important role in facilitating genome annotation. We are not alone in this view. Indeed, structure prediction and structure modeling (i.e. homology modeling) are quickly becoming a routine part of many protein analyses and proteome annotation efforts (6). Annotation systems such as BASYS (4), BACMAP (5), PEDANT (7) and others all depend on large-scale secondary structure predictions to assist in identifying possible functions, to determine subcellular locations or to identify structural genomics targets.
Beyond its application to routine annotation, structure prediction can also be used to assess organism-specific trends in secondary structure content, to identify protein folds, to identify domains, and to estimate the proportion of ‘unfolded’ or unstructured proteins in a given genome (8–10). It is also common to use structure predictions or structure modeling to decide where and how to subclone protein fragments for expression, where to join or insert gene fragments, or where to add affinity tags for protein purification. It is also possible to use secondary structure prediction to calibrate circular dichroism (CD) and Fourier transform infrared spectroscopy (FTIR) measurements when monitoring the folding or unfolding proteins with no known 3D structure (11).
Over the past decade, a number of excellent structure prediction and structure modeling servers have emerged. These include Porter (12) and PsiPred (13) for secondary structure prediction of soluble proteins, SWISS-MODEL (14) and 3D JigSaw (15) for homology modeling, TMHMM (16) for transmembrane helix prediction, Pred-TMBB for transmembrane β-barrel prediction (17) and SignalP (18) for signal peptide prediction. However, most of these tools are highly specialized, single application servers that perform only one type of prediction, for just one sequence at a time. Consequently, if a newly sequenced protein does not fit neatly into one of the standard prediction categories it is difficult to get a very complete or well-annotated result. For instance, if a protein (such as OmpA) happens to have a signal peptide, an N-terminal membrane spanning domain, and a C-terminal soluble cytoplasmic domain that is homologous to a known 3D structure, a user may have to visit at least four different web servers to get a complete structural analysis of the protein. Trying to merge these disparate results into a single, coherent prediction would require a significant amount of manual inspection, reformatting and alignment. If one wished to analyze hundreds of proteins of a similar nature, such a task would prove to be very challenging, especially given the fact that very few structure prediction tools support local installations. Indeed, even fewer are distributed as open source applications. As a result, the integration, local installation or customization of these tools is almost impossible.
Another limitation to essentially all secondary structure prediction systems is the fact that they do not fully exploit the information that is already available in the protein structure databases (i.e. the PDB). We have recently shown that by finding sequence homologues in the PDB and by using a process called 3D-to-2D mapping, it is possible to increase the accuracy of secondary structure prediction (of soluble proteins) by as much as 10% (19). A similar approach has recently been applied to Porter as a means of significantly improving its secondary structure predictions (20). Applying this simple structure mapping protocol to predicting the structure of transmembrane helix or transmembrane β-barrel, proteins could potentially improve their corresponding prediction accuracies by a similar amount.
In an effort to address some of the current shortcomings in structure prediction and structure-based annotation, we have developed the PROTEUS2 structure prediction server. PROTEUS2 is unique among structure prediction servers in that it bundles signal peptide identification, transmembrane helix prediction, transmembrane β-strand prediction, (soluble) secondary structure prediction and homology modeling (i.e. 3D structure generation) into a unified prediction pipeline that supports both single sequence and large, multi-sequence submissions. Using a combination of machine learning and database comparison techniques, PROTEUS2 is able to achieve very high levels of predictive accuracy for signal peptide identification (Q2 = 95%), membrane spanning regions (Q2 = 87%) and secondary structure (Q3 = 81%). It is also capable of producing high-quality 3D models (with downloadable coordinates) when appropriate database matches are found. The PROTEUS2 server is freely available, as is the source code and binaries for a stand-alone (nonserver) version.
PROGRAM DESCRIPTION
PROTEUS2 is composed of two parts, a front-end web-interface (written in Perl and HTML) and a back-end consisting of five different structure prediction programs (written in Java, Perl and C/C++) along with four local databases (about 310 Mbytes in size). The front-end accepts both FASTA and raw sequence data. The sequences may be either pasted or typed into the text box or uploaded through a file browse button. The server accepts both single sequence and multiple sequence files. As part of the server interface, users must select the kingdom to which the source organism belongs (Gram+, Gram− and Eukaryote) to improve the quality of the signal peptide predictions. For multi-sequence submissions users must provide an email address to which the results can be sent.
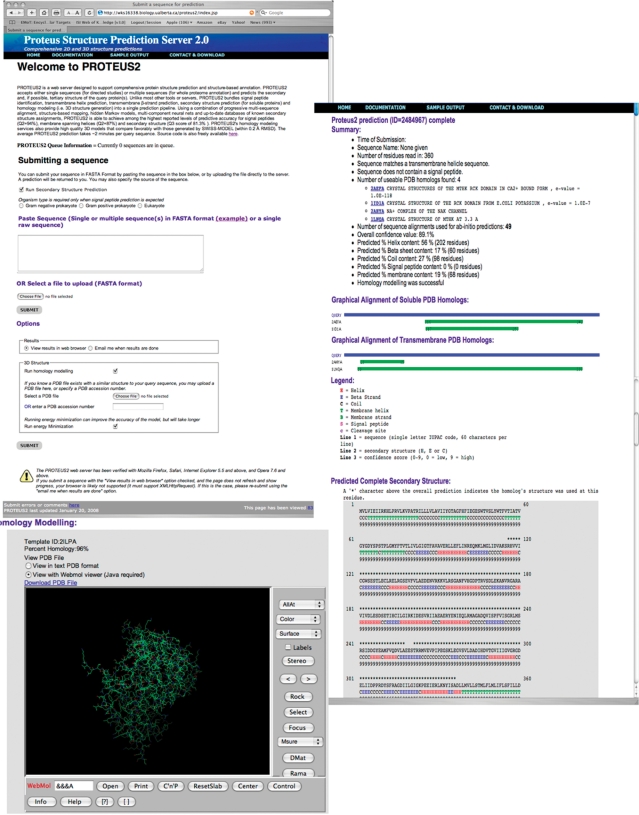
The output for a typical PROTEUS2 prediction consists of several pages of hyperlinked or scrollable text files (Figure 1) including sequence/structure alignments, predictions for signal peptide location and cleavage sites, membrane spanning regions (both helices and β-strands) and putative or known domains. Signal peptide segments are marked with an ‘S’, membrane spanning helices are identified with a ‘T’, membrane β-strands are identified with a ‘B’, regular helices are marked with an ‘H’, regular β-strands with an ‘E’, coil regions with a ‘C’ and signal peptide cleavage sites with a lowercase ‘c’. PROTEUS2 also generates confidence scores for each type of secondary structure (additional details about the confidence scores are available on PROTEUS2 documentation pages). If a 3D structure is generated, the PDB coordinates, information about the matching PDB structure, the predicted alignment, the sequence identity, the number of modeled residues and a hyperlink to view the resulting structure through the WebMol viewer (21) are provided. Users may override PROTEUS2 default choice of structure templates by preselecting a PDB file under the PROTEUS2 options menu. It is also possible to toggle the energy minimization option on or off to improve either the quality or speed of structure generation.
Figure 1.
A screenshot montage illustrating the typical output from a PROTEUS2 prediction of the structure of the Ca2+-gated K+ channel from Methanobacterium autotrophicum. Shown are examples of the membrane spanning, soluble/cytoplasmic secondary structure predictions and homology models generated from its analysis.
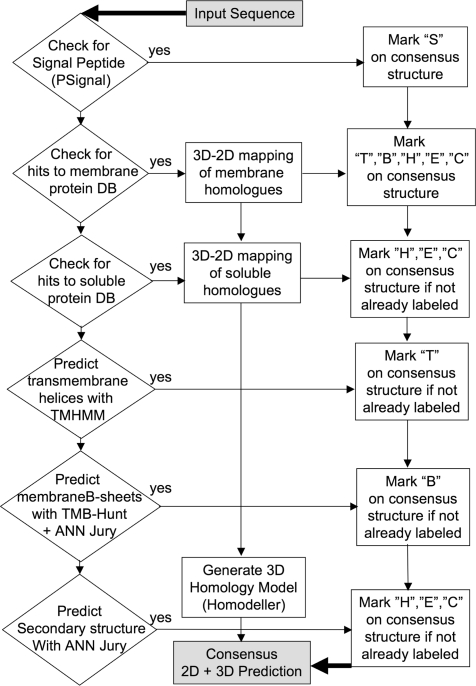
In order to perform its structure predictions most accurately and efficiently, PROTEUS2 follows a strict hierarchy of analyses and database comparisons (see flow chart in Figure 2). When a query sequence is received, PROTEUS2 initially performs a signal peptide identification step using a profile-based hidden Markov modeling (HMM). This step is followed by BLASTing the query protein against a database of 2587 proteins with known signal peptides obtained from the PPT-DB (22). If a significant (Expect <10−10) match is found, the resulting alignment is used to transfer the known signal peptide data of the template molecule to the query protein using a technique called 3D-to-2D mapping (19). If a signal peptide (and cleavage site) is identified, the sequence is truncated and sent to the next prediction step. If no signal peptide is identified, the server keeps the query sequence unchanged. In the next step, the query sequence is aligned against a specially constructed database of 275 solved helical membrane proteins. This membrane helix database was also obtained from the PPT-DB. If a significant (Expect <10−10) match is found, the resulting alignment is used to transfer the known secondary structure of the template membrane molecule to the query protein using the previously mentioned 3D-to-2D mapping technique. If no match can be found, the program applies TMHMM 2.0 (16) to predict the membrane spanning helical regions. To reduce the number of false positives identified by TMHMM, the query sequence is also BLASTed against the PPT-DB's cytoplasmic protein database (16 618 nonredundant, nonmembrane proteins obtained from the PDB) and any significant matches (Expect <10−10) have their previously identified membrane helices removed. If no membrane helices can be found, a similar protocol is used to determine whether the query protein has any transmembrane β-barrel segments. In particular, PROTEUS2 first uses TMB-HUNT (23) to identify whether the query sequence is a potential membrane β-barrel protein. Once the existence of a membrane β-barrel protein has been confirmed, the location of the β-strands is predicted using an approach that combines a locally developed secondary structure predictor (19) with a 3D-to-2D mapping of the β-strands from homologous membrane β-barrel proteins.
Figure 2.
A flow chart showing the logic and sequential operations performed by PROTEUS2 on each query sequence.
After the signal peptides and transmembrane segments (helices or β-strands) have been identified, PROTEUS2 proceeds to the third structure prediction step. In this step, PROTEUS2 initially compares the query sequence to PPT-DB's collection of water-soluble proteins of known structure. The secondary structures for these proteins were assigned using VADAR as reported previously (19,21). If a significant (Expect <10−10) match is found, the same 3D-to-2D mapping procedure is used to assign or predict the secondary structure. If no match can be found, the PROTEUS2 applies a locally developed ‘jury of experts’ prediction method to predict the secondary structures as described in our earlier publication (19).
Once the signal peptide, membrane spanning regions and secondary structures have been predicted, the query sequence is directed to a locally developed homology modeling program called HOMODELLER (24). If one or more PDB files with a BLAST expect score of <10−10 was previously found in either of the earlier 3D-to-2D mapping steps, the highest scoring homolog covering the largest region of the query sequence is used as a template for HOMODELLER to build a 3D structure. While PROTEUS2 always succeeds in generating linear (i.e. secondary structure) predictions for any query sequence, 3D structures will only be generated if the query protein passes the HOMODELLER thresholds.
ALGORITHMS AND TESTING
In developing the HMMs for signal peptide prediction program (called PredictSP), more than 2000 signal peptides and 1200 control peptides (covering the N-terminal sequence to the cleavage site) were obtained from the SignalP data set (24). These test sequences are partitioned into groups belonging to Gram+ (453 peptides), Gram− (1199 peptides) and Eukaryotic (1599 peptides) organisms. The predictor was trained to recognize not only the existence of a signal peptide, but also its length and cleavage site. PredictSP was subject to 10-fold cross-validation during the testing and training phases. The performance of PredictSP was further enhanced by employing previously developed 3D-to-2D mapping methods using the signal peptide database (SPDB) obtained from the PPT-DB.
PROTEUS2 transmembrane prediction program builds from two previously developed and freely available programs, TMHMM (16) and TMB-HUNT (23). TMHMM uses a HMM to identify transmembrane helices, while TMB-HUNT uses amino acid composition statistics to identify potential membrane β-barrel proteins. We coupled TMB-HUNT to a locally developed secondary structure predictor (19) to obtain ‘de novo’ secondary structure assignments of the transmembrane β-strands. The performance of both transmembrane prediction methods was enhanced by exploiting the 3D-to-2D mapping methods that we previously developed for predicting the secondary structure of water-soluble proteins (19). Training and testing of the algorithms was conducted using the relevant transmembrane PPT-DB databases (22).
In implementing the ‘jury of experts’ approach for predicting the secondary structure of water-soluble proteins, we used the same programs and methods we developed previously (19). In training and testing the program nearly 2000 sequence-unique sequences were analyzed, requiring some 100 hours of CPU time. The program was written such that secondary structure predictions from the consensus predictor could be overridden if a homologous protein could be found in PROTEUS2 secondary structure database.
PROTEUS2 homology modeling module, HOMODELLER, is quite conventional and employs standard homology modeling techniques (14,15). It uses BLAST to search through an internal, nonredundant version of the PDB database (which is updated monthly) to find and align the closest matching sequence homolog. Mismatched residues in the template sequence are changed to match the query sequence, but with the same χ1-angles of the template. Gaps in coil regions are handled using a loop library (consisting of >13 000 loops derived from high-resolution structures in the PDB). The inserted regions are superimposed and then iteratively adjusted to fit the surrounding regions using a cyclic coordinate descent algorithm (25). The resulting structure is energy minimized using a locally developed torsion angle minimizer, called GAfolder. GAfolder uses cyclic coordinate decent in combination with a simple genetic algorithm to perform conformational sampling. The energy function, also developed locally, uses a knowledge-based potential that includes threading energies, hydrogen bond energies, van der Waals interactions and other components. Additional details about the potential function and the algorithm used in GAfolder are provided in PROTEUS2 documentation pages. The method has been extensively tested and refined using hundreds of known structures and 1000s of decoys. More recently, HOMODELLER was used to model more than 100 000 protein structures for the BacMap project (5).
RESULTS AND EVALUATION
All of the algorithms and programs employed in PROTEUS2 have been extensively tested and used, both internally and externally (some for as long as 6 years). As with any structure prediction suite, a key measure of its utility is its predictive accuracy. The accuracy of each PROTEUS2 program employing machine-learning methods was tested in several ways, including accuracy during the training/testing phases, accuracy using newly acquired or ‘unseen’ data and accuracy as measured by independent evaluation tools such as EVA and TMH-Benchmark (26,27). The results of these evaluations along with comparisons to other tools or servers are listed in Table 1. Additional details about the benchmarking protocols, the evaluation methods, the programs tested along with the test data sets themselves are provided in PROTEUS2 web ‘Documentation’ page, under the heading ‘How PROTEUS2 Measures Up’ Web (Table 1a–g).
Table 1.
Summary of PROTEUS2 structure prediction performance relative to other structure prediction tools
| Signal peptide prediction performance (PPT-DB SPDB test set) | ||
|---|---|---|
| Program or Server | Q2 (Gram-) (%) | Q2 (Gram+) (%) |
| PROTEUS2 | 95 | 94 |
| SubLoc | 91 | 86 |
| SignalP(3.0) | 96 | 97 |
| Transmembrane helix prediction performance (TMH Benchmark test set) | ||
| Program or Server | Q2 (%) | # False positives |
| PROTEUS2 | 91 | 0 |
| TMHMM | 80 | 1 |
| HMMTOP | 80 | 6 |
| DAS | 72 | 16 |
| Transmembrane helix prediction performance (PPT-DB-TMH test set) | ||
| Program or Server | Q2 (%) | # False neg. (missed prots) |
| PROTEUS2 | 87 | 0 |
| TMHMM | 82 | 8 |
| Transmembrane β-barrel detection performance (PPT-DB ‘All’ protein data set) | ||
| Program or Server | Q2 (%) | Accuracy (TMB versus glob) (%) |
| PROTEUS2 | 100 | 100 |
| TMB-Hunt | 78 | 99 |
| Transmembrane β-strand prediction performance (PPT-DB -TMB test set) | ||
| Program or Server | Q2 (%) | |
| PROTEUS2 | 86 | |
| Pred-TMBB | 73 | |
| Non-membrane secondary structure prediction performance (EVA test set) | ||
| Program or Server | Q3 | SOV |
| PROTEUS2 | 81 | 82 |
| Porter | 77 | 76 |
| JNET | 72 | 73 |
| PSIPred | 77 | 78 |
| Non-membrane secondary structure prediction performance (test set of 125) | ||
| Program or Server | Q3 | SOV |
| PROTEUS2 | 88 | 90 |
| Porter | 76 | 81 |
| JNET | 73 | 77 |
| PHD | 76 | 78 |
| Homology modeling performance | ||
| Program or Server | RMSD all (Å) | RMSD backbone (Å) |
| PROTEUS2 | 1.83 | 0.99 |
| Swiss-Model | 1.62 | 0.86 |
| 3D JigSaw | 1.94 | 0.97 |
Details of the test sets and test conditions are given in the text.
For signal peptide, transmembrane helix and transmembrane β-barrel predictions we evaluated both the per-residue prediction accuracy (Q2) as well as the ability of the predictors to correctly identify proteins with or without these structural features (sensitivity/specificity). Secondary structure predictions of soluble proteins were evaluated using only the Q3 and segment overlap (SOV) scores. The results in Table 1 report the performance of PROTEUS2 combined predictors. However, since each of the four 1D predictors used both de novo predictions and homology-based methods we also assessed: (i) the performance of the de novo predictors alone; (ii) the performance of the homology-based structure predictors alone and (iii) the performance of the combined predictors. When measuring the performance of any 3D-to-2D mapping prediction the standard approach is to iteratively remove each sequence from the database and to perform the prediction with that sequence (19–21). This prevents one from simply predicting the structure of the query protein using the query itself. It is also important to report the per-residue accuracy as well as the coverage (the percentage of query proteins that returned an answer). These results along with additional information about prediction sensitivity/specificity are shown in Tables 2–5 of PROTEUS2 ‘Documentation’ webpage, under the ‘How PROTEUS2 Measures Up’ heading.
To assess PROTEUS2 signal peptide prediction accuracy, a data set of 2587 complete protein sequences with experimentally confirmed signal peptides as well as a data set of 16 618 cytoplasmic proteins (with no signal peptides in their sequence) was extracted from the PPT-DB. The signal peptide set included proteins from each of the three major classes of organisms (Gram+, Gram− and Eukaryote). PROTEUS2 was compared against SubLoc and SignalP 3.0 (using their default values) measuring both sensitivity/specificity and per-residue prediction accuracy (Q2). As seen in Table 1, our predictor performs nearly as well as SignalP 3.0 and somewhat better than SubLoc, with Q2 scores of 95% for Gram−, 94% for Gram+ signal peptides. This compares favorably to SignalP 3.0 Q2 scores of 96% for Gram−, 97% for Gram+ signal peptides.
To assess transmembrane helix prediction accuracy, two tests were employed. In one, the PROTEUS was assessed against the 2247 proteins (globular and transmembrane) used in TMH-Benchmark (27). In the other, a data set of 275 complete protein sequences with experimentally confirmed transmembrane helices was extracted from the PPT-DB. From the TMH-Benchmark results, PROTEUS2 was able to achieve a Q2 score of 91% (for high-resolution structures), which is 11% better than any other method. It also had the best performance in distinguishing between globular proteins and membrane proteins (0 false positives). Because there is some uncertainty in the transmembrane assignments for some of TMH Benchmarks high-resolution data set, we performed a second evaluation using the experimentally confirmed data set derived from PPT-DB. In this assessment, we compared the performance of PROTEUS2 to TMHMM only. As seen in Table 1, PROTEUS2 does significantly better than TMHMM in both Q2 scores (87% versus 81%) and in globular/membrane confusion scores (0 false negatives versus 8 false negatives).
The assessment of transmembrane β-barrel detection and β-sheet prediction was done using a set of experimentally determined 49 transmembrane β-barrel and 16 618 water-soluble, globular proteins obtained from PPT-DB. The program was compared against TMB-HUNT and Pred-TMBB (using their default parameters). As can be seen from Table 1, the sensitivity/specificity in distinguishing globular proteins from transmembrane β-barrel proteins for PROTEUS2 is particular good (sp = sn = 100%) versus sp = 78%, sn = 99% for TMB-HUNT. Likewise PROTEUS2 Q2 scores for transmembrane β-sheet prediction, thanks to the use of 3D-to-2D mapping methods, are also very high (86% versus 73%).
The assessment of PROTEUS2 performance on globular proteins or nonmembrane secondary structure prediction was done using two approaches: (i) through a ‘blind’ test and comparison on the latest EVA training set (1644 sequence-unique proteins) and (ii) through analysis of 125 randomly chosen proteins that were recently solved by X-ray and NMR. The latter set was chosen to simulate a more realistic case of predicting the secondary structure of sequences found in a proteome (which tend not to be sequence-unique). In both cases, the Q3 and SOV scores were calculated for each protein in the test sets. Both sets are also available from PROTEUS2 Download page. Results were compared to Porter (12), PSIPred (13), PHD (6) and JNET (28). As seen in Table 1, the results are essentially identical to those reported previously, with PROTEUS2 performing somewhat better than other globular protein structure prediction servers (Q3 of 81–88% versus Q3 of 72–77%).
An assessment of PROTEUS2 performance for homology modeling was also performed. In one case, 37 proteins with sequence identities ranging from 21.2% to 99.2% (the PDB IDs are listed on the PROTEUS2 documentation pages) were modeled using PROTEUS2 and 3D JigSaw (using default parameters). In the second case, 33 proteins with similar sequence identity ranges were modeled using PROTEUS2 and SWISS-MODEL (also using default parameters). In each case, identical template structures were used for the pairwise comparisons. The resulting structures were compared using a variety of criteria including backbone RMSD, all-atom RMSD, percentage of torsion angle violations and a variety of energy terms (average hydrogen bond energies, threading energies, bump scores). The results are summarized briefly in Table 1 and in more detail in on the PROTEUS2 website (Tables 6 and 7 of the web ‘Documentation’ page). The SWISS-MODEL structures had average backbone RMSDs of 0.86 Å and all-atom RMSDs of 1.62 Å relative to the ‘correct’ or known structure. The 3D-JigSaw structures had average backbone RMSDs of 0.97 Å and all-atom RMSDs of 1.94 Å relative to the ‘correct’ or known structure. The PROTEUS2 structures had average backbone RMSDs of 0.99 Å and all-atom RMSDs of 1.83 Å (for the SWISS-MODEL set) and average backbone RMSDs of 1.04 Å and all-atom RMSDs of 2.00 Å (for the 3D JigSaw set).
These comparisons show that the HOMODELLER structures are essentially comparable to those generated by 3D-JigSaw and SWISS-MODEL. While there are a growing number of homology modeling servers with increasingly impressive capabilities (29,30) it is important to point out that PROTEUS2 is not just a homology modeling server and that it is designed to provide considerably more information about a protein or proteins of interest, regardless of whether a homology model can be generated or not. Furthermore, because PROTEUS2 is uniquely configured to handle multiple sequences we believe it should fill a unique niche for the structural genomics or structural proteomics community.
CONCLUSION
PROTEUS2 is an integrated web server that makes use of robust machine learning techniques, in-house modeling programs and an extensive collection of customized structural databases to provide both comprehensive and highly accurate protein structure predictions. The target audience or target users for PROTEUS2 are structural biologists and scientists working in structural genomics or structural proteomics projects where something ‘structural’ has to be known about the proteins prior to being selected, mutated, truncated, engineered, cloned or expressed. Essentially, the role of the PROTEUS2 server is to facilitate target selection, target structure determination and structure-based protein/proteome annotation. It is important to emphasize that PROTEUS2 is not a ‘meta-server’, in that it does not depend on external servers to perform its predictions. Almost all of the software and databases used in PROTEUS2 were developed and tested locally, meaning that the code (and databases) may be easily ported to other sites or platforms. Overall, we believe the open source nature of the PROTEUS2 software, the high level of accuracy of PROTEUS2 linear predictors along with PROTEUS2 3D structure generation capabilities could make it a very useful addition to the current arsenal of structure prediction tools available to both protein chemists and bioinformaticians.
ACKNOWLEDGEMENTS
Funding for this project was provided by the Alberta Prion Research Institute, PrioNet, NSERC and Genome Alberta. Funding to pay the Open Access publication charges for this article was provided by the Alberta Prion Research Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hall N. Advanced sequencing technologies and their wider impact in microbiology. J. Exp. Biol. 2007;210:1518–1525. doi: 10.1242/jeb.001370. [DOI] [PubMed] [Google Scholar]
- 2.Riley M, Abe T, Arnaud MB, Berlyn MK, Blattner FR, Chaudhuri RR, Glasner JD, Horiuchi T, Keseler IM, Kosuge T, et al. Escherichia coli K-12: a cooperatively developed annotation snapshot—2005. Nucleic Acids Res. 2006;34:1–9. doi: 10.1093/nar/gkj405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stothard P, Wishart DS. Automated bacterial genome analysis and annotation. Curr. Opin. Microbiol. 2006;9:505–510. doi: 10.1016/j.mib.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Van Domselaar GH, Stothard P, Shrivastava S, Cruz JA, Guo A, Dong X, Lu P, Szafron D, Greiner R, Wishart DS. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res. 2005;33(Web Server issue):W455–W459. doi: 10.1093/nar/gki593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stothard P, Van Domselaar G, Shrivastava S, Guo A, O’Neill B, Cruz J, Ellison M, Wishart DS. BacMap: an interactive picture atlas of annotated bacterial genomes. Nucleic Acids Res. 2005;33(Database issue):D317–D320. doi: 10.1093/nar/gki075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32(Web Server issue):W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mewes HW, Frishman D, Mayer KF, Munsterkotter M, Noubibou O, Pagel P, Rattei T, Oesterheld M, Ruepp A, Stumpflen V. MIPS: analysis and annotation of proteins from whole genomes in 2005. Nucleic Acids Res. 2006;34(Database issue):D169–D172. doi: 10.1093/nar/gkj148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter P, Liu J, Rost B. PEP: predictions for entire proteomes. Nucleic Acids Res. 2003;31:410–413. doi: 10.1093/nar/gkg102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Rost B. Comparing function and structure between entire proteomes. Protein Sci. 2001;10:1970–1979. doi: 10.1110/ps.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oldfield CJ, Cheng Y, Cortese MS, Brown CJ, Uversky VN, Dunker AK. Comparing and combining predictors of mostly disordered proteins. Biochemistry. 2005;44:1989–2000. doi: 10.1021/bi047993o. [DOI] [PubMed] [Google Scholar]
- 11.Ullman CG, Haris PI, Smith KF, Sim RB, Emery VC, Perkins SJ. Beta-sheet secondary structure of an LDL receptor domain from complement factor I by consensus structure predictions and spectroscopy. FEBS Lett. 1995;371:199–203. doi: 10.1016/0014-5793(95)00916-w. [DOI] [PubMed] [Google Scholar]
- 12.Pollastri G, McLysaght A. Porter: a new, accurate server for protein secondary structure prediction. Bioinformatics. 2005;21:1719–1720. doi: 10.1093/bioinformatics/bti203. [DOI] [PubMed] [Google Scholar]
- 13.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 14.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bates PA, Kelley LA, MacCallum RM, Sternberg MJE. Enhancement of protein modelling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Proteins. 2001;(Suppl 5):39–46. doi: 10.1002/prot.1168. [DOI] [PubMed] [Google Scholar]
- 16.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 17.Bagos PG, Liakopoulos TD, Spyropoulos IC, Hamodrakas SJ. PRED-TMBB: a web server for predicting the topology of beta-barrel outer membrane proteins. Nucleic Acids Res. 2004;32(Web Server issue):W400–W404. doi: 10.1093/nar/gkh417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 19.Montgomerie S, Sundararaj S, Gallin WJ, Wishart DS. Improving the accuracy of protein secondary structure prediction using structural alignment. BMC Bioinform. 2006;7:301. doi: 10.1186/1471-2105-7-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollastri G, Martin AJ, Mooney C, Vullo A. Accurate prediction of protein secondary structure and solvent accessibility by consensus combiners of sequence and structure information. BMC Bioinform. 2007;8:201. doi: 10.1186/1471-2105-8-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walther D. WebMol—a Java-based PDB viewer. Trends Biochem. Sci. 1997;22:274–275. doi: 10.1016/s0968-0004(97)89047-0. [DOI] [PubMed] [Google Scholar]
- 22.Wishart DS, Arndt D, Berjanskii M, Guo AC, Shi Y, Shrivastava S, Zhou J, Zhou Y, Lin G. PPT-DB: the protein property prediction and testing database. Nucleic Acids Res. 2008;36(Database issue):D222–D229. doi: 10.1093/nar/gkm800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garrow AG, Agnew A, Westhead DR. TMB-Hunt: a web server to screen sequence sets for transmembrane beta-barrel proteins. Nucleic Acids Res. 2005;33(Web Server issue):W188–W192. doi: 10.1093/nar/gki384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wishart DS, Case DA. Use of chemical shifts in macromolecular structure determination. Methods Enzymol. 2001;338:3–34. doi: 10.1016/s0076-6879(02)38214-4. [DOI] [PubMed] [Google Scholar]
- 25.Canutescu AA, Dunbrack R.L., Jr. Cyclic coordinate descent: a robotics algorithm for protein loop closure. Protein Sci. 2003;12:963–972. doi: 10.1110/ps.0242703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eyrich VA, Marti-Renom MA, Przybylski D, Madhusudhan MS, Fiser A, Pazos F, Valencia A, Sali A, Rost B. EVA: continuous automatic evaluation of protein structure prediction servers. Bioinformatics. 2001;17:1242–1243. doi: 10.1093/bioinformatics/17.12.1242. [DOI] [PubMed] [Google Scholar]
- 27.Kernytsky A, Rost B. Static benchmarking of membrane helix predictions. Nucleic Acids Res. 2003;31:3642–3654. doi: 10.1093/nar/gkg532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuff JA, Barton GJ. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 29.Wallner B, Elofsson A. All are not equal: a benchmark of different homology modeling programs. Protein Sci. 2005;14:1315–1327. doi: 10.1110/ps.041253405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nayeem A, Sitkoff D, Krystek S., Jr A comparative study of available software for high-accuracy homology modeling: from sequence alignments to structural models. Protein Sci. 2006;15:808–824. doi: 10.1110/ps.051892906. [DOI] [PMC free article] [PubMed] [Google Scholar]