Abstract
RNA interference (RNAi) with small interfering RNA (siRNA) has become a powerful tool in functional and medical genomic research through directed post-transcriptional gene silencing. In order to apply RNAi technique for eukaryotic organisms, where frequent alternative splicing results in diversification of mRNAs and finally of proteins, we need spliced mRNA isoform silencing to study the function of individual proteins. AsiDesigner is a web-based siRNA design software system, which provides siRNA design capability to account for alternative splicing for mRNA level gene silencing. It provides numerous novel functions including the designing of common siRNAs for the silencing of more than two mRNAs simultaneously, a scoring scheme to evaluate the performance of designed siRNAs by adopting currently known key design factors, a stepwise off-target searching with BLAST and FASTA algorithms and checking the folding secondary structure energy of siRNAs. To do this, we developed a novel algorithm to evaluate the common target region, where siRNAs can be designed to knockdown a specific mRNA isoform or more than two mRNA isoforms from a target gene simultaneously. The developed algorithm and the AsiDesigner were tested and validated as very effective throughout widely performed gene silencing experiments. It is expected that AsiDesigner will play an important role in functional genomics, drug discovery and other molecular biological research. AsiDesigner is freely accessible at http://sysbio.kribb.re.kr/AsiDesigner/.
INTRODUCTION
Short interfering RNAs (siRNAs) degrade specific target mRNAs, the transcription products of genes through the RNA interference (RNAi) mechanism. The importance of siRNAs has been widely recognized and a series of studies on siRNA applications have been conducted in diverse research fields including functional genomics and drug discovery (1,2).
Various siRNA design algorithms and programs have been developed and serviced to provide good performance in siRNA design for biologists (3–26). Traditionally, many of them have considered certain rules to evaluate the siRNA efficacy, such as Tuschl's rules (27–29) containing several parameters: 3′overhangs, GC contents, absence of internal repeats, homology to other mRNAs, absence of SNPs and RNA secondary structure, etc. The binding energy factors of double-stranded region were noticed and many researches suggested that thermodynamic properties of siRNA, including asymmetry for RISC assembly, play a critical role in determining the functions of siRNAs (6,8,9,30–32). Reynold et al. (7), Mückstein et al. (33) and Choi et al.'s (34) work also considered relative binding energy profiles for rational siRNA design. The performance of published siRNA efficacy predictors were compared and assessed by several groups (25,35,36), particularly, Matveeva et al. (25) exhaustively assessed several algorithms by statistical analysis with the published experimental data and derived a new efficient method predicting siRNA efficiency for in their recent work.
These algorithms are known to be efficient for siRNA selection; however, they are limited to gene-level silencing. Restated, their knock-down capacity is limited to a specific mRNA or to all the isoforms from a target gene. For eukaryotic organisms, alternative splicing contributes to the diversity of gene functions resulting in the production of multiple proteins from a gene (37). Thus, it is necessary to control an individual spliced mRNA isoform producing a specific protein to study the functions of alternatively splicing genes completely (38). Moreover, it is feasible to knockdown a specific combination of more than two mRNA isoforms simultaneously from a gene. In this case, the possible combination of targets, dependent on the purpose of research and the number of combinations, will be too numerous to prepare siRNA libraries for all the occasions. Thus, a new algorithm and tool to design common siRNAs for a given combination of targets considering alternative splicing will be useful in this field.
We developed a novel algorithm to evaluate a common target region where siRNAs can be designed to knockdown a specific mRNA isoform or multiple mRNA isoforms from a target gene. AsiDesigner is a web-based siRNA design software system providing siRNA design capability to take into account alternative splicing for mRNA level gene silencing. It provides many useful functions, which include designing common siRNAs for silencing more than two mRNAs simultaneously, a scoring scheme to evaluate the performance of designed siRNAs by adopting currently known key design factors, a two-step off-target searching with sequence alignment algorithms and checking the folding secondary structure energy of siRNAs.
METHODS
Evaluation of a common target region
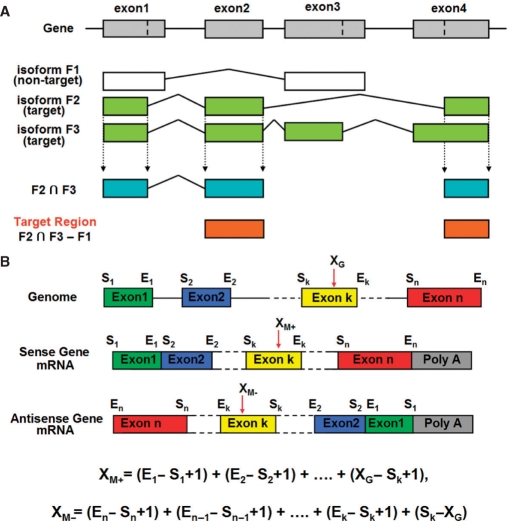
AsiDesigner uses a novel and effective algorithm for selecting siRNA target regions, which considers alternative splicing. The purpose of this algorithm is to evaluate a region to design siRNAs targeting one or more mRNAs selectively from an available mRNA isoform set of a target gene. To design siRNAs, a target region must be selected from common exons of the targeted mRNA isoforms, and any exons of non-target mRNA isoforms should be excluded from this target region. Therefore, as shown in Figure 1A, the final target region will be equivalent to the subtraction of the union of non-target isoform mRNA exons from the intersection of target isoform mRNA exons. There could be two methods to achieve this purpose: the first may use the result of multiple sequence alignment of all the available mRNA isoforms and the second may use the result of mapping all the available isoform mRNAs on the genome. In this algorithm, we applied the latter method and the mapping information of mRNAs on the genome of UCSC Genome Bioinformatics (http://genome.ucsc.edu/) was utilized.
Figure 1.
Schematic diagrams for evaluating a target region to design siRNAs for specific alternatively spliced isoforms (A) and conversion process of a siRNA position on the genome, XG to the mRNA-based coordinate, XM+ from sense strand coded gene or XM− from antisense strand coded gene (B).
In an actual evaluation of target region, this process is carried out with the information of both target and non-target exon positions. First, common target exon regions are evaluated as follows:
Select one target isoform mRNA as the main target and other target mRNA isoforms as the co-targets.
Evaluate common exon region(s) of the main target and one of the co-targets by comparing their positions on the genome.
Evaluate common exon region(s) of previously evaluated common exon region(s) and one of the other co-targets.
Repeat (ii) and (iii) until all co-targets are processed.
Next, the final target region evaluation is similar as the preceding method. It is accomplished by the repeated exclusion of the overlapping regions of common target exon regions with the non-target exon regions.
The target template sequence for siRNA design can be extracted from genome or mRNA sequence corresponding to the target region. However, the actual target of siRNA is mRNA and there may be a little difference between the genome and mRNA sequences by post-transcriptional processing such as RNA editing. Since we need to use a mRNA sequence to extract templates, the position information of target region on the genome was converted to that on the main target mRNA by the Equation 1 for a sense-strand coded gene on the genome when the target region is on the k-th exon as shown in Figure 1B.
| 1 |
Here, XM+ is the position on the main target mRNA corresponding to XG, the position on the genome, for a gene made of n exons. Si is the starting position of i-th exon and Ei is the end position of i-th exon. The k is the index for the exon including the target region of the siRNA. If a gene is coded in an antisense strand on the genome, the Equation 2 may be used.
| 2 |
Rules for siRNA selection and efficiency scoring
After evaluating the common target region and sequence, actual siRNA design is performed by the optimized siRNA design algorithm implementing relative binding energy profile discrimination method published previously (34). The siRNA candidates generated from the target region were screened by applying the 5 selection rules and the efficiencies of the candidate siRNAs were evaluated by the performance scoring rules.
siRNA selection rules
Candidate siRNA sequences are screened or selected based on the following selection rules:
G/C content: users set up allowable range of GC content (default: 30–62%).
Maximum consecutive bases (default: maximum 5 for A or T, maximum 3 for G or C and maximum 7 for G/C mixed).
Existence of single nucleotide polymorphism (SNP) in the siRNA target region.
Self-alignment energy of siRNA (ΔG < −4 kcal/mol by the Mfold).
Maximum identical bases to the non-target mRNAs for off-target filtering (default: equal to or less than 15/19).
Scoring rules for siRNA efficiency
The performance scores are evaluated by an optimized scoring scheme of AsiDesigner to assess the efficiencies of siRNA candidates that passed the prior selection criteria. This performance scoring system assigns individual scores (Zi) for five key design parameters affecting the performance of siRNAs and combines them linearly with the statistically evaluated weighting factors (Wi) for each design parameter to get the final total performance score (Zt) (34).
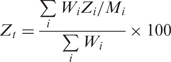 |
3 |
where i is an index for each design parameter: 36–53% GC content, number of A/Us at 15–19th bases from 5′ sense position, G/C existence at the first 5′ sense position, A/U existence at the 19th base from 5′ sense position, and the relative binding energy profile, respectively, and W1 = 0.11, W2 = 0.07, W3 = 0.15, W4 = 0.19, and W5 = 0.90. Individual score of design parameter Zi comes from the scoring scheme for each design parameter and Mi is the maximum value of individual score: Z1 is 10 when the content of G/C ranges from 36% to 53% and 0 when it does not belong within the range; Z2 is the number of A/Us in 5 bases of 3′end; Z3 is 1 when the base of 5′ end is G/C or 0 when it isn't; Z4 is 1 when the base of 3′ end is A/U or 0 when it isn't; Z5 is the score calculated with the relative binding energy profile (34). The respective value of Mi is as follows: M1 = 10, M2 = 5, M3 = 1, M4 = 1 and M5 = 100.
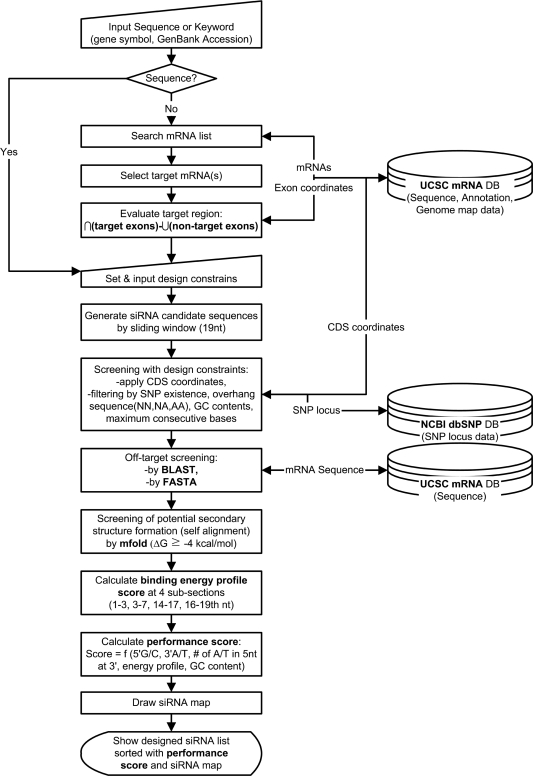
Weighting factor Wi for each design parameter was decided as a combination of the values that maximize the t-statistics of the total performance score Zt between the highly efficient siRNA group and inefficient siRNA group in the training set from Khvorova et al.'s (6) work. The final design parameter, the relative binding energy profile score was evaluated by optimized scoring scheme based on the performance asymmetry of siRNAs with the relative binding energy profile difference between the highly efficient and inefficient siRNA groups (34). A schematic diagram for a siRNA design process in AsiDesigner is shown in Figure 2.
Figure 2.
Schematic diagram of a data processing flow for siRNA design in AsiDesigner.
IMPLEMENTATION
AsiDesigner was developed as a web application program based on Java, JSP/JSF technologies and MySQL relational database. It has been serviced by a Linux machine with eight CPUs of 2.2 GHz using Apache web server with Tomcat web container. Users may access and use AsiDesigner under a user-friendly GUI environment interactively through the internet.
Input and options
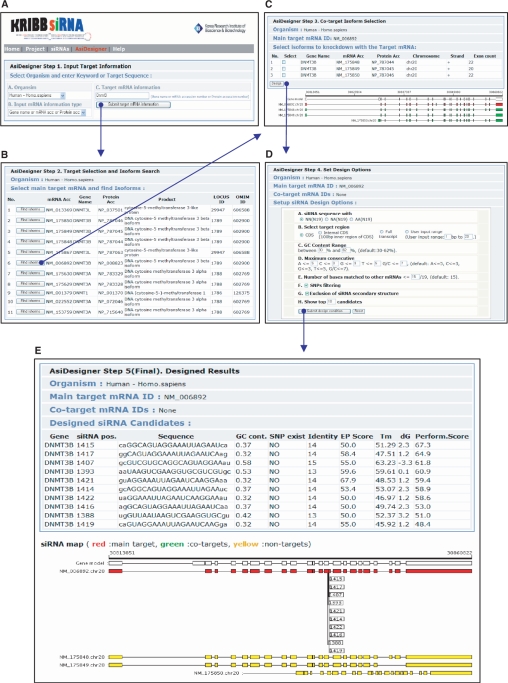
With a step-by-step input interface (Figure 3), users can design siRNAs by AsiDesigner with ease. After searching and selecting target mRNA(s) or submitting a target mRNA sequence, AsiDesigner, with a number of design options, designs and provides a sequential list of optimized siRNAs by using the internal algorithm and scoring scheme.
Figure 3.
Screenshots for the siRNA design procedure in AsiDesigner: step 1 for submitting target information in terms of gene symbol or GenBank accession number of mRNA or protein (A), step 2 for target mRNA selection and available isoform search (B), step 3 for co-target isoform selection (C), step 4 for setting siRNA design options (D) and step 5 for the output of designed siRNA list (E).
To search and select target mRNA(s), users undertake the following steps (Figure 3A–C):
Selection of organism and retrieving target gene (mRNAs).
Selection of main target mRNA from the retrieved mRNA list and searching available mRNA isoforms from the target gene.
Selection of co-target mRNA(s) from the retrieved available mRNA isoforms.
In input window (Figure 3A), an organism list box for human, mouse and rat is provided as choices. The organism information chosen is then used for searching genes or mRNAs, off-target filtering and SNP checking for the selected organism. In the next step, in order to search for the target gene or mRNA information, users may use keywords, i.e. gene symbols (for example ‘BCL2L1’ or ‘BRCA2’) or GenBank accession numbers of mRNAs or proteins (for example ‘NM_138578’ or ‘NP_612815’), which is the keyword-based input mode. Also, it is possible to use DNA or RNA sequence as an input in FASTA format alternatively if users have only partial sequence of target mRNA, which is the sequence-based input mode.
After submitting target gene information in the keyword-based input mode, AsiDesigner retrieves gene information from the mRNA database corresponding to the given keyword and print out a matched mRNA list (Figure 3B). Users check whether the mRNA from the target exists in the list and select one of them as a main target, then all the available mRNA isoforms of the main target mRNA are retrieved and listed in a table with a map (Figure 3C). After checking co-target mRNAs in the available mRNA isoform list, a window for setting up design options appears (Figure 3D). In the design option window, users can set options such as types of overhang (AA, NA or NN), sub-target region, range of GC content, identity limit for off-target filtering, SNP-based filtering, secondary structure-based filtering and the number of siRNAs to be shown. Detailed information with respect to siRNA design with AsiDesigner is provided on the web page.
In the sequence-based input mode, once a user submits a sequence after selecting organism, a design option window appears and the following processes will be identical to the keyword-based input mode. Input target sequence should be in FASTA format and should include a string of unmodified RNA/DNA bases, i.e. A, U/T, G and C only. Any other characters in the sequence will be edited and removed. Multiple FASTA format sequences are not supported; only the first sequence will be processed. The maximum length of the input sequence is 5 kb.
Output
After setting and submitting the design options, AsiDesigner designs optimized siRNA candidates by the internal algorithm and the performance scoring scheme. AsiDesigner provides a sequential list of the designed siRNAs by their evaluated performance scores. The output includes the position on the main target mRNA, siRNA sequence, GC content, existence of SNP site in the region, identity from off-target search, energy-profile score and estimated melting temperature. Users may check and compare the position of the designed siRNAs in the output map that shows the exon structure of the target gene and non-target mRNA isoforms (Figure 3E). Sequences of designed siRNAs are provided as the form of 23 bp RNA sequences: the left 1–2nd bases lower cased are the overhang of the anti-sense siRNA, the next 3–21st bases upper cased are the double stranded region, and the last 22–23rd bases lower cased are the overhang of the sense siRNA.
Data and used tools
For the keyword-based gene search, genic annotation and genome map (physical position) data of mRNAs (hg18, March 2006; mm8, March 2006; rn4, November 2004) from UCSC Genome Bioinformatics were parsed and stored in MySQL relational database (RDBMS). Exon coordinate information of 24 375 mRNAs from 18 392 human genes, 20 162 mRNAs from 18 722 mouse genes and 10 152 mRNAs from 9977 rat genes were stored in Gene information and mRNA sequence database. To perform off-target search, NCBI BLAST (39) and FASTA (40) programs were utilized with RefSeq mRNA sequence set (Release 22 March 2007) from UCSC Genome Bioinformatics. Mfold (41) program was used for evaluating folding energies in the secondary structures of designed siRNAs. To test SNP sites existence in designed siRNAs, SNP data from NCBI dbSNP (Build 127, March 2007) were used with MySQL RDBMS.
Performance of designed siRNAs
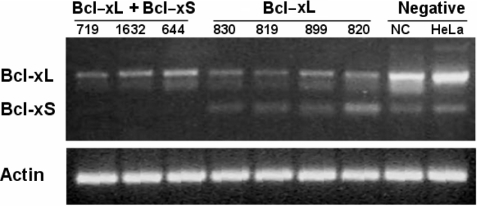
To see the capability of AsiDesigner on isoform specific siRNA design, silencing experiments of the human BCLX (BCL2L1) gene were conducted with two isoform mRNAs, Bcl-xL (NM_138578) and Bcl-xS (NM_001191) as shown in Figure 4. Three siRNAs to knockdown both isoform mRNAs (siRNA set 1: 719, 1632 and 644) and four siRNAs to knockdown Bcl-xL mRNA alone (siRNA set 2: 830, 819, 899 and 820) were designed and transfected into the human HeLa cell line. Knockdown rates were measured by RT-PCR and northern blotting analysis. In case of siRNA set 1, the average knockdown rates for Bcl-xL and Bcl-xS were 73 and 100%, respectively, and 71 and 2%, respectively, for siRNA set 2. Consequently we verified that the siRNA set 2 targeting Bcl-xL alone silenced Bcl-xL properly, while siRNA set 1 targeting both isoforms knocked down both Bcl-xL and Bcl-xS as expected.
Figure 4.
Results of gene silencing experiments for two alternative isoforms, Bcl-xL (NM_138578) and Bcl-xS (NM_001191) of human BCLX (BCL2L1) gene by RT–PCR. siRNA transfection was performed into the human cell line (HeLa) for three siRNAs to knock-down both isoform mRNAs (siRNA set 1: 719, 1632 and 644) and four siRNAs to knockdown Bcl-xL mRNA alone (siRNA set 2: 830, 819, 899 and 820). The average knockdown rates for Bcl-xL and Bcl-xS were 73 and 100%, respectively, by siRNA set 1 and 71 and 2%, respectively, by siRNA set 2.
Besides this result, the performance of the optimized scoring scheme for siRNA efficacy adopted in AsiDesigner was validated through numerous gene silencing experiments, a total of over 300 siRNAs from five gene families (NFKB, STE kinase, TK kinase, Caspase and CUT; Supplementary Table S1 to Table S6) and BIRC5 gene. Also, it was used to design 67 siRNAs from seven genes (EGFP, MAPT, SNCA, SNCB, ACTB, HSP22 and GARS) with our collaborative researches and also to build predesigned human kinase and phosphatase siRNA libraries. Most of gene silencing experiments were carried out with effective siRNAs and the accuracy of siRNA efficacy prediction based on 315 effective siRNAs from five gene families and three ineffective siRNAs from BIRC5 gene was 0.79 based on 75% knockdown rate (Table S1).
FUTURE DEVELOPMENTS
Since BLAST algorithm for off-target screening is not sufficient (42,43), we applied FASTA program for siRNA candidates that passed from BLAST search in the AsiDesigner, which is very time consuming. We plan to adapt miRNA-like off-target filtering algorithm for nonreal-time-based siRNA design using email system in the future (44,45).
CONCLUSION
AsiDesigner provides users with a useful novel function with which to design siRNAs for multiple target mRNAs considering alternative splicing. Candidate siRNA sequences based on the currently known key design factors and many advanced techniques are expected to have optimized efficiencies. Through numerous gene silencing experiments undertaken by several different groups, the AsiDesigner performance has been validated to be very efficient; we anticipate that AsiDesigner may be used effectively for various functional genomics, drug discovery, as well as for other molecular biology studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to the numerous anonymous AsiDesigner reviewers for their comments. We also thank many researchers for their participation in the massive experimental validation of siRNA performance. This research was supported by the Cerebrovascular Disease Project of the Korean Ministry of Science & Technology (MOST) and in part by the Korean HapMap Project. D.L. was supported by the Ministry of Knowledge Economy, Korea, under the ITRC supervised by the IITA (IITA-2008-C1090-0801-0001). Funding to pay the Open Access publication charges for this article was provided by MOST.
Conflict of interest statement. None declared.
REFERENCES
- 1.Couzin J. Breakthrough of the year: small RNAs make big splash. Science. 2002;298:2296–2297. doi: 10.1126/science.298.5602.2296. [DOI] [PubMed] [Google Scholar]
- 2.Dorsett Y, Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- 3.Birmingham A, Anderson E, Sullivan K, Reynolds A, Boese Q, Leake D, Karpilow J, Khvorova A. A protocol for designing siRNAs with high functionality and specificity. Nat. Protocols. 2007;2:2068–2078. doi: 10.1038/nprot.2007.278. [DOI] [PubMed] [Google Scholar]
- 4.Chalk AM, Wahlestedt C, Sonnhammer EL. Improved and automated prediction of effective siRNA. Biochem. Biophys. Res. Commun. 2004;319:264–274. doi: 10.1016/j.bbrc.2004.04.181. [DOI] [PubMed] [Google Scholar]
- 5.Yuan B, Latek R, Hossbach M, Tuschl T, Lewitter F. siRNA Selection Server: an automated siRNA oligonucleotide prediction server. Nucleic Acids Res. 2004;32(Web Server issue):W130–W134. doi: 10.1093/nar/gkh366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 7.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 8.Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 9.Amarzguioui M, Prydz H. An algorithm for selection of functional siRNA sequences. Biochem. Biophys. Res. Commun. 2004;316:1050–1058. doi: 10.1016/j.bbrc.2004.02.157. [DOI] [PubMed] [Google Scholar]
- 10.Sætrom P. Predicting the efficacy of short oligonucleotides in antisense and RNAi experiments with boosted genetic programming. Bioinformatics. 2004;20:3055–3063. doi: 10.1093/bioinformatics/bth364. [DOI] [PubMed] [Google Scholar]
- 11.Ui-Tei K, Naito Y, Takahashi F, Haraguchi T, Ohki-Hamazaki H, Juni A, Ueda R, Saigo K. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huesken D, Lange J, Mickanin C, Weiler J, Asselbergs F, Warner J, Meloon B, Engel S, Rosenberg A, Cohen D, et al. Design of a genome-wide siRNA library using an artificial neural network. Nat. Biotechnol. 2005;23:995–1001. doi: 10.1038/nbt1118. [DOI] [PubMed] [Google Scholar]
- 13.Jagla B, Aulner N, Nelly PD, Song D, Volchuk A, Zatorski A, Shum D, Mayer T, De Angelis DA, Ouerfelli O, et al. Sequence characteristics of functional siRNAs. RNA. 2005;11:864–872. doi: 10.1261/rna.7275905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shabalina SA, Spiridonov AN, OgurtsovA Y. Computational models with thermodynamic and composition features improve siRNA design. BMC Bioinform. 2005;7:65. doi: 10.1186/1471-2105-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vert JP, Foveau N, Lajaunie C, Vandenbrouck Y. An accurate and interpretable model for siRNA efficacy prediction. BMC Bioinform. 2006;7:520. doi: 10.1186/1471-2105-7-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henschel A, Buchholz F, Habermann B. DEQOR: a web-based tool for the design and quality control of siRNAs. Nucleic Acids Res. 2004;32:W113–W120. doi: 10.1093/nar/gkh408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong D, Ferrell J.E., Jr. Picking a winner: new mechanistic insights into the design of effective siRNAs. Trends Biotechnol. 2004;22:451–454. doi: 10.1016/j.tibtech.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 18.Yiu SM, Wong PW, Lam TW, Mui YC, Kung HF, Lin M, Cheung YT. Filtering of ineffective siRNAs and improved siRNA design tool. Bioinformatics. 2005;21:144–151. doi: 10.1093/bioinformatics/bth498. [DOI] [PubMed] [Google Scholar]
- 19.Ge G, Wong GW, Luo B. Prediction of siRNA knockdown efficiency using artificial neural network models. Biochem. Biophys. Res. Commun. 2005;336:723–728. doi: 10.1016/j.bbrc.2005.08.147. [DOI] [PubMed] [Google Scholar]
- 20.Levenkova N, Gu Q, Rux JJ. Gene specific siRNA selector. Bioinformatics. 2004;20:430–432. doi: 10.1093/bioinformatics/btg437. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Mu FY. A web-based design center for vector-based siRNA and siRNA cassette. Bioinformatics. 2004;20:1818–1820. doi: 10.1093/bioinformatics/bth164. [DOI] [PubMed] [Google Scholar]
- 22.Naito Y, Yamada T, Ui-Tei K, Morishita S, Saigo K. siDirect: highly effective, target-specific siRNA design software for mammalian RNA interference. Nucleic Acids Res. 2004;32:W124–W129. doi: 10.1093/nar/gkh442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santoyo J, Vaquerizas JM, Dopazo J. Highly specific and accurate selection of siRNAs for high-throughput functional assays. Bioinformatics. 2005;21:1376–1382. doi: 10.1093/bioinformatics/bti196. [DOI] [PubMed] [Google Scholar]
- 24.Arziman Z, Horn T, Boutros M. E-RNAi: a web application to design optimized RNAi constructs. Nucleic Acids Res. 2005;33:W582–W588. doi: 10.1093/nar/gki468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matveeva O, Nechipurenko Y, Rossi L, Moore B, Sætrom P, Ogurtsov AY, Atkins JF, Shabalina SA. Comparison of approaches for rational siRNA design leading to a new efficient and transparent method. Nucleic Acids Res. 2007;35:e63. doi: 10.1093/nar/gkm088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holen T. Efficient prediction of siRNAs with siRNArules 1.0: an open-source JAVA approach to siRNA algorithms. RNA. 2006;12:1620–1625. doi: 10.1261/rna.81006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in mammalian cell culture. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 28.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21 and 22 nt RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 31.Lin SL, Chang D, Ying SY. Asymmetry of intronic pre-miRNA structures in functional RISC assembly. Gene. 2005;356:32–38. doi: 10.1016/j.gene.2005.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hutvagner G. Small RNA asymmetry in RNAi: function in RISC assembly and gene regulation. FEBS Lett. 2005;579:5850–5857. doi: 10.1016/j.febslet.2005.08.071. [DOI] [PubMed] [Google Scholar]
- 33.Mückstein U, Tafer H, Hackermüller J, Bernhart SH, Stadler PF, Hofacker IL. Thermodynamics of RNA-RNA binding. Bioinformatics. 2006;22:1177–1182. doi: 10.1093/bioinformatics/btl024. [DOI] [PubMed] [Google Scholar]
- 34.Choi Y, Park H, Choung S, Kim Y, Kim S, Park S, Kim S, Yoon G, Choi K, Kang H. 2005. Patent PCT/KR2005/004207 (WO 2006/062369) [Google Scholar]
- 35.Saetrom P, Snøve O., Jr. A comparison of siRNA efficacy predictors. Biochem. Biophys. Res. Commun. 2004;321:247–253. doi: 10.1016/j.bbrc.2004.06.116. [DOI] [PubMed] [Google Scholar]
- 36.Boese Q, Leake D, Reynolds A, Read S, Scaringe SA, Marshall WS, Khvorova A. Mechanistic insights aid computational short interfering RNA design. Methods Enzymol. 2005;392:73–96. doi: 10.1016/S0076-6879(04)92005-8. [DOI] [PubMed] [Google Scholar]
- 37.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 38.Celotto AM, Graveley BR. Exon-specific RNAi: a tool for dissecting the functional relevance of alternative splicing. RNA. 2002;8:718–724. doi: 10.1017/s1355838202021064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 40.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1998;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 42.Snøve O, Holen T. Many commonly used siRNAs risk off-target activity. Biochem. Biophys. Res. Commun. 2004;319:256–263. doi: 10.1016/j.bbrc.2004.04.175. [DOI] [PubMed] [Google Scholar]
- 43.Yamada T, Morishita S. Accelerated off-target search algorithm for siRNA. Bioinformatics. 2005;21:1316–1324. doi: 10.1093/bioinformatics/bti155. [DOI] [PubMed] [Google Scholar]
- 44.Brown K, Samarsky D. RNAi off-targeting: light at the end of the tunnel. J. RNAi Gene Silencing. 2006;2:175–177. [PMC free article] [PubMed] [Google Scholar]
- 45.Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.