Abstract
Although mass spectrometry has been frequently used to identify proteins, there are no web servers that provide comprehensive functional annotation of those identified proteins. It is necessary to provide such web service due to a rapid increase in the data. We, therefore, introduce MassNet, which provides (i) physico-chemical analysis information, (ii) KEGG pathway assignment (iii) Gene Ontology mapping and (iv) protein–protein interaction (PPI) prediction for the data from MASCOT, Prospector and Profound. MassNet provides the prediction information for PPIs using both 3D structural interaction and experimental interaction deposited in PSIMAP, BIND, DIP, HPRD, IntAct, MINT, CYGD and BioGrid. The web service is freely available at http://massnet.kr or http://sequenceome.kobic.re.kr/MassNet/.
INTRODUCTION
Mass spectrometry (MS) is the key method for proteomics (1). MS is widely used to study complex cellular proteomes and low abundance proteins (1–3). With it we can rapidly identify proteins and obtain information for protein complexes and posttranslational modification (3). MS data are used to produce genome-scale data (4). Presently, the functional annotation of MS data often requires researchers to navigate numerous web-accessible primary data servers. In order to analyze large-scale data, one approach is to provide access to an integrated web server that contains rich bio-information with graphic interfaces (5). Several MS data processing systems have been developed to handle these challenges. They are MASCOT (http://www.matrixscience.com) (2), Prospector (http://prospector.ucsf.edu) and Profound (http://prowl.rockefeller.edu) (6). These systems provide protein identification data using public databases such as SwissProt (http://www.ebi.ac.uk/swissprot) and NCBInr (http://www.ncbi.nlm.nih.gov). These web services do not include the functional annotation of MS data and do not supply the latest version of the analysis tools. To provide an easy and automated pipeline for functional annotation of given MS results, we constructed a web-based server, MassNet. The use of MassNet does not require any application installation and it is easy to use.
METHODS
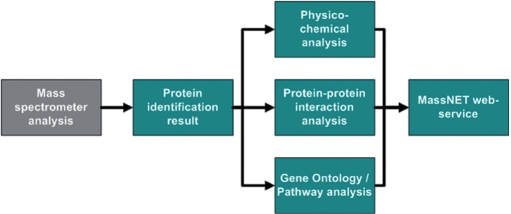
To analyze MS data, various protein annotation resources are required. Therefore, we integrated major protein sequence databases, protein–protein interaction (PPI) databases, Gene Ontology (GO) (http://www.geneontology.org) (7), KEGG pathway (http://www.genome.jp/kegg) (8) and bioinformatics analysis tools such as SignalP. This system has four major parts: (i) a nonredundant protein database, (ii) a physico-chemical property analysis module, (iii) a function annotation module and (iv) a PPI prediction module. A schematic workflow of MassNet is shown in Figure 1.
Figure 1.
The schematic workflow of MassNet.
Construction of the nonredundant protein database
In order to identify proteins from MS data, researchers use various protein sequence databases such as NCBInr, SwissProt and trEMBL. However, there can be confusion among protein identifiers. Because of this problem, all protein identifiers were relationally linked. We integrated the protein sequence databases (Swiss-Prot, trEMBL, NCBInr, RefSeq, Ensembl and IPI) using only perfect-matching sequences. The database unifies protein IDs of the same sequence, summarizes annotations and descriptions of proteins from a range of organisms representing all three major kingdoms of life: eukaryotes, prokaryotes and viruses. Therefore, the root identifier (Sequenceome_ID) can contain several protein identifiers from all available databases. The Sequenceome_ID database is a nonredundant sequence database of 6 856 434 proteins (April 2008).
Analysis of physico-chemical properties of proteins
The physico-chemical properties of MS data are important to understand biological functions. Especially, the prediction of hydropathy and subcelular localization of MS data is closely related to find membrane proteins which are involved in cellular processes and protein classes as drug targets (9). We used modules from Biopython (http://biopython.org) (10) to calculate hydropathy profile, GRAVY score (the average hydropathy score for all the amino acids), protein length, molecular weight, amino acid distribution, isoelectric point and protein instability index (11). For the subcellular localization prediction, we predicted transmembrane helices and signal peptides using Phobius (http://phobius.sbc.su.se) (12) and SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP) (13) programs. In order to provide physico-chemical information without any time delay, we provide precalculated physico-chemical properties for all nonredundant protein sequences. Whole proteins’ physico-chemical properties are also provided as summary tables or figures. If a set of proteins was input, the user can acquire information on the protein set's physico-chemical distribution against whole-protein distribution of the organism. If the identified protein set was from the membrane fraction of an organism, the user compares the relative transmembrane protein abundances between the organism's whole-protein set and the identified protein set. Therefore, this summary information can be used to evaluate the input data quality.
Integration of annotation information
MassNet provides biological function information by using KEGG pathways and GO. The KEGG pathway database and GO represent an attempt to assign known proteins into known biological pathways and are updated regularly (8). MassNet assigns proteins to KEGG pathways thorough ID mapping and shows color-coded proteins in the context of biochemical pathway maps using KEGG API. In order to find significant associations of GO terms with queried proteins, we assigned proteins into GO categories and GO-slim (14) through ID mapping. In order to gain more accurate statistical test results of KEGG and GO assignment, we added Fisher's exact test algorithm (P-value).
Prediction of PPI
The prediction of PPI is based on PSIMAP (protein structural interactome MAP) (http://psimap.com, http://psibase.kobic.re.kr) (15,16) and PEIMAP (protein experimental interactome MAP) (17). The basic algorithm of PSIMAP infers interactions among proteins by using their homologs. Interactions among domains or proteins for known PDB (Protein Data Bank) (http://www.rcsb.org/pdb) structures are the basis of the predictions. If an unknown protein has a homolog to a domain, PSIMAP assumes that the query tends to interact with its homolog's partners. Its concept is called ‘homologous interaction’ (18–20). The original interaction between two proteins or domains is based on the Euclidean distance. Therefore, PSIMAP gives a structure-based interaction prediction (15). On the other hand, PEIMAP is a well-established method that uses public resources of experimentally confirmed protein interaction information such as BIND (http://bond.unleashedinformatics.com) (21), DIP (http://dip.doe-mbi.ucla.edu) (22), IntAct (http://www.ebi.ac.uk/intact) (23), MINT (http://mint.bio.uniroma2.it/mint) (24), HPRD (http://www.hprd.org) (25), CYGD (http://mips.gsf.de/genre/proj/yeast) (26) and BioGrid (http://www.thebiogrid.org) (27). We constructed a nonredundant PPI database from the source databases. We carried out a redundancy check to remove identical protein sequences from the source interaction databases using PERL (http://www.perl.org). Now, it contains 116 773 proteins and 229 799 interactions. The accuracy of PEIMAP is dependent on the confidence of each resource. In order to reduce the false positive rate of PEIMAP, we computed the final ‘combined score’ for each pair of proteins which were predicted by PEIMAP and PSIMAP algorithms. This scoring methodology has been proposed by published articles including the STRING server (http://string.embl.de) (28). Users can easily predict PPI for queried proteins in a list and can examine PPIs with a network viewer.
USER INTERFACE
Input
The query interface allows the user to submit an HTML file from the mass spectrometry or a TAB-delimited text file. The tab-delimited file must contain protein names in the first column. Detailed information about the TAB-delimited file format is described on the ‘HOW TO USE’ page. MassNet can use four types of MS data formats, i.e. MASCOT, Prospector, Profound and TAB-delimited file.
Output
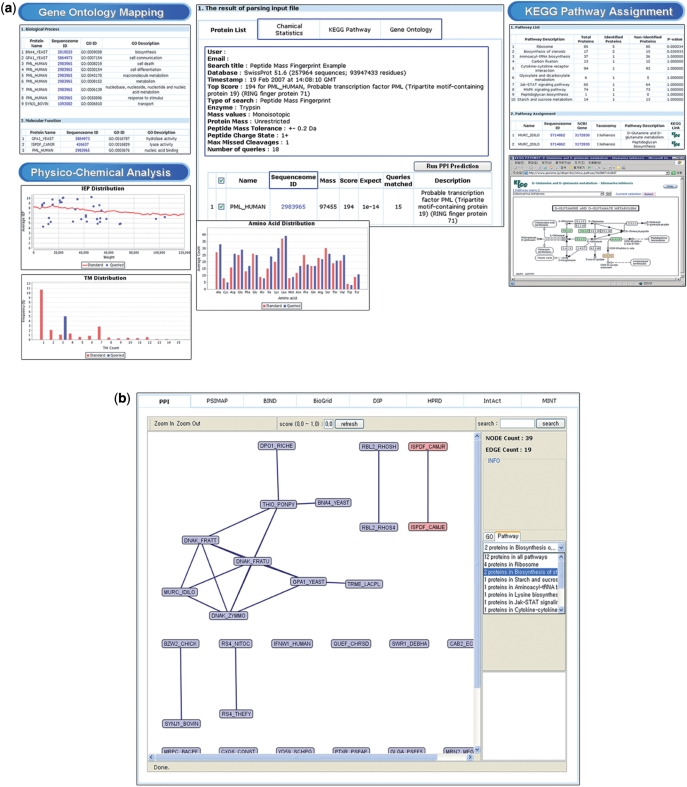
After uploading the query file, users can obtain the annotation information as in Figure 2a. The annotation results consist of five parts: (i) a protein list page, (ii) the physico-chemical property of each protein, (iii) a PPI prediction page, (iv) a KEGG pathway page and (v) a GO page.
Figure 2.
Screenshots of MassNet annotation results. (a) Panel in the middle is the protein list table. KEGG Pathway tab shows KEGG pathway assignment and metabolic pathway graph (right panels). Gene Ontology tab shows proteins assigned to GO categories (left top panel). Chemical Statistics tab shows the input protein set's physico-chemical distribution against whole protein distribution of the organism (left bottom panels). (b) Protein-protein interactions of user-selected proteins are visualized by a network viewer. Rectangular shapes are protein nodes. The black connecting lines indicate interactions among the nodes. The two red rectangular nodes are proteins that are selected by the users through the right hand side panel. When users select the right pull down menus in the right panel, the left drawing canvas shows highlighted protein nodes.
The protein list page shows a table describing protein names and scores, which are parsed from the query file. The KEGG pathway and the GO pages show the number of proteins, which belong to the categories of KEGG pathways and GO. By clicking the ‘Run PPI Prediction’ button at the top of the protein list table, the user can acquire the PPI information for selected proteins. The PPI page shows PEIMAP and PSIMAP (see Methods section) data at two separated tables.
By clicking Sequenceome_IDs at all pages, users can access two pages, i.e. a Same IDs page and a Chemical Property page. The same IDs page shows the identical sequences at various protein sequence databases and provides the hyperlinks to original database web pages. In order to provide clear information, MassNet provides a viewer for PPI networks as in Figure 2b.
IMPLEMENTATION
The MassNet web server runs on a Linux server. It combines a MySQL (http://www.mysql.com) database with a dynamic web interface using Java Server Pages (http://java.sun.com/products/jsp). Data preprocessing is implemented in Perl and Python, and the network viewer for PPI was constructed using Java.
CONCLUSION
The functional analysis and interpretation of the large-scale MS data are still a challenging task. An automatic approach is necessary for tens of thousands of MS data collected throughout the world. MassNet is the first web server that provides various kinds of functional information, such as physico-chemical properties, biological pathways, gene ontology and PPI, for MS data. MassNet is easy to use and provides information through an automatic annotation for queried proteins.
ACKNOWLEDGEMENTS
We thank Maryana Bhak for editing the article and Suan Cho for making the figures. This work was supported by a grant from the KRIBB Research Initiative Program of Korea, by the Korea Science and Engineering Foundation (KOSEF) grant funded by the Korean government (MOST) (No. M10508040002-07N0804-00210) and supported by a Korea Basic Science Institute K-Mep grant (T28021) to J-.S.C. Funding to pay the Open Access publication charges for this article was provided by MOST (No. M10508040002-07N0804-00210).
Conflict of interest statement. None declared.
REFERENCES
- 1.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 3.Pandey A, Mann M. Proteomics to study genes and genomes. Nature. 2000;405:837–846. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- 4.Kemmeren P, van Berkum NL, Vilo J, Bijma T, Donders R, Brazma A, Holstege FC. Protein interaction verification and functional annotation by integrated analysis of genome-scale data. Mol. Cell. 2002;9:1133–1143. doi: 10.1016/s1097-2765(02)00531-2. [DOI] [PubMed] [Google Scholar]
- 5.Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome biology. 2003;4:P3. [PubMed] [Google Scholar]
- 6.Eng JK, McCormack AL, Yates J.RIII. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 7.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 10.Chapman B, Chang J. Biopython: python tools for computational biology. ACM SIGBIO Newsletter. 2000;20:15–19. [Google Scholar]
- 11.Guruprasad K, Reddy B.VB, Pandit MW. Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 1990;4:155–161. doi: 10.1093/protein/4.2.155. [DOI] [PubMed] [Google Scholar]
- 12.Kall L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 13.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protocols. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 14.Camon E, Magrane M, Barrell D, Lee V, Dimmer E, Maslen J, Binns D, Harte N, Lopez R, Apweiler R. The Gene Ontology Annotation (GOA) Database: sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 2004;32:D262–D266. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park J, Lappe M, Teichmann SA. Mapping protein family interactions: intramolecular and intermolecular protein family interaction repertoires in the PDB and yeast. J. Mol. Biol. 2001;307:929–938. doi: 10.1006/jmbi.2001.4526. [DOI] [PubMed] [Google Scholar]
- 16.Gong S, Yoon G, Jang I, Bolser D, Dafas P, Schroeder M, Choi H, Cho Y, Han K, Lee S, et al. PSIbase: a database of protein structural interactome map (PSIMAP) Bioinformatics. 2005;21:2541–2543. doi: 10.1093/bioinformatics/bti366. [DOI] [PubMed] [Google Scholar]
- 17.Kim JG, Park D, Kim BC, Cho SW, Kim YT, Park YJ, Cho HJ, Park H, Kim KB, Yoon KO, et al. Predicting the interactome of Xanthomonas oryzae pathovar oryzae for target selection and DB service. BMC Bioinform. 2008;9:41. doi: 10.1186/1471-2105-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marcotte EM, Pellegrini M, Thompson MJ, Yeates TO, Eisenberg D. A combined algorithm for genome-wide prediction of protein function. Nature. 1999;402:83–86. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- 19.Walhout AJ, Sordella R, Lu X, Hartley JL, Temple GF, Brasch MA, Thierry-Mieg N, Vidal M. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science. 2000;287:116–122. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- 20.Deane CM, Salwinski L, Xenarios I, Eisenberg D. Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol. Cell Proteomics. 2002;1:349–356. doi: 10.1074/mcp.m100037-mcp200. [DOI] [PubMed] [Google Scholar]
- 21.Bader GD, Hogue CW. BIND—a data specification for storing and describing biomolecular interactions, molecular complexes and pathways. Bioinformatics. 2000;16:465–477. doi: 10.1093/bioinformatics/16.5.465. [DOI] [PubMed] [Google Scholar]
- 22.Xenarios I, Rice DW, Salwinski L, Baron MK, Marcotte EM, Eisenberg D. DIP: the database of interacting proteins. Nucleic Acids Res. 2000;28:289–291. doi: 10.1093/nar/28.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hermjakob H, Montecchi-Palazzi L, Lewington C, Mudali S, Kerrien S, Orchard S, Vingron M, Roechert B, Roepstorff P, Valencia A, et al. IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004;32:D452–D455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zanzoni A, Montecchi-Palazzi L, Quondam M, Ausiello G, Helmer-Citterich M, Cesareni G. MINT: a Molecular INTeraction database. FEBS Lett. 2002;513:135–140. doi: 10.1016/s0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]
- 25.Peri S, Navarro JD, Kristiansen TZ, Amanchy R, Surendranath V, Muthusamy B, Gandhi TK, Chandrika KN, Deshpande N, Suresh S, et al. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 2004;32:D497–D501. doi: 10.1093/nar/gkh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guldener U, Munsterkotter M, Kastenmuller G, Strack N, van Helden J, Lemer C, Richelles J, Wodak SJ, Garcia-Martinez J, Perez-Ortin JE, et al. CYGD: the comprehensive yeast genome database. Nucleic Acids Res. 2005;33:D364–D368. doi: 10.1093/nar/gki053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]