Abstract
We demonstrate the feasibility of generating thousands of transgenic Drosophila melanogaster lines in which the expression of an exogenous gene is reproducibly directed to distinct small subsets of cells in the adult brain. We expect the expression patterns produced by the collection of 5,000 lines that we are currently generating to encompass all neurons in the brain in a variety of intersecting patterns. Overlapping 3-kb DNA fragments from the flanking noncoding and intronic regions of genes thought to have patterned expression in the adult brain were inserted into a defined genomic location by site-specific recombination. These fragments were then assayed for their ability to function as transcriptional enhancers in conjunction with a synthetic core promoter designed to work with a wide variety of enhancer types. An analysis of 44 fragments from four genes found that >80% drive expression patterns in the brain; the observed patterns were, on average, comprised of <100 cells. Our results suggest that the D. melanogaster genome contains >50,000 enhancers and that multiple enhancers drive distinct subsets of expression of a gene in each tissue and developmental stage. We expect that these lines will be valuable tools for neuroanatomy as well as for the elucidation of neuronal circuits and information flow in the fly brain.
Keywords: enhancer, gene expression, promoter, transcription, transgenic
The functional elements of the nervous system and the neuronal circuits that process information are not genes but cells. Consequently, the classic genetic methods that have been so powerful in elucidating embryonic development and other processes in Drosophila melanogaster are not adequate to probe the function of the nervous system (1). Instead, we will need to be able to assay and manipulate the function of individual neurons with the same facility as we can now assay and manipulate the function of individual genes.
A variety of genetically encoded probes have been developed that allow researchers to visualize individual neurons to study anatomy, as well as to monitor and modulate the activity of neurons to study physiology and behavior. The utility of these probes is highly dependent on the precision with which their expression can be directed to small subsets of neurons in reproducible, controllable, and convenient ways. The primary objective of the work described in this report was to expand the tools available to accomplish such precise, controlled expression in the nervous system of D. melanogaster.
Researchers have known for more than 20 years how to identify and, to some extent, manipulate the promoters and enhancers that control the temporal and spatial expression of individual genes in Drosophila (2). This work, and similar studies in other animals, has revealed that the complex spatial and temporal expression pattern of a gene usually results from the combined action of a set of individual enhancer elements that act, in a largely autonomous manner, to dictate aspects of the expression of that gene (3, 4). The number of enhancers per gene varies widely but is generally thought to be in the range of 2 to 10 in Drosophila (5).
Because individual enhancers appear to represent the fundamental cis-acting modules through which gene expression patterns are generated, our objective was to identify a large set of enhancers that could each reproducibly drive expression of a reporter gene in a distinct, small subset of cells in the adult CNS. Ideally, the number of defined expression patterns should be large enough that, in sum, they would cover the entire brain several times over in a variety of overlapping patterns.
The feasibility of this approach depends on a number of factors. First, enhancers from a wide range of genes whose core promoters contain different sequence motifs must each function robustly when placed in a defined genomic location with a common core promoter. Second, the expression pattern driven by a given enhancer must be highly reproducible from animal to animal. Third, the expression patterns driven by individual enhancers should contain an appropriately small fraction of the cells in the brain to make them useful tools for neuroanatomy and behavioral genetics. Finally, the methods for transgenesis and for identifying suitable enhancers must be efficient enough to permit the generation of the required thousands of transgenic lines. Here, we report the development of a strategy that we believe meets all four of these criteria.
Results and Discussion
Overview of Experimental Strategy.
We selected 925 genes for which available expression data or predicted function implied expression in a subset of cells in the adult brain, for example, genes encoding transcription factors, neuropeptides, ion channels, transporters, and receptors [supporting information (SI) Dataset S1]. We spanned the flanking upstream and downstream intergenic regions of these genes, as well as any of their introns larger than 300 bp, with fragments of DNA that averaged 3 kb in length and overlapped (in regions that could not be covered by a single fragment) by ≈1 kb. The fragments were generated by PCR from genomic DNA using primers designed to lie in areas of low evolutionary conservation to minimize disruption of individual enhancers. Because the average size of an enhancer element is only a few hundred base pairs (5), we expected that nearly all enhancers would be intact in at least one fragment. This process generated 5,200 fragments that were cloned, sequence-verified, and inserted upstream of a core promoter (Fig. 1A). In ≈200 cases in which the upstream intergenic region was small, we generated PCR fragments that also contained the start site of transcription and used them to create transcriptional fusion constructs.
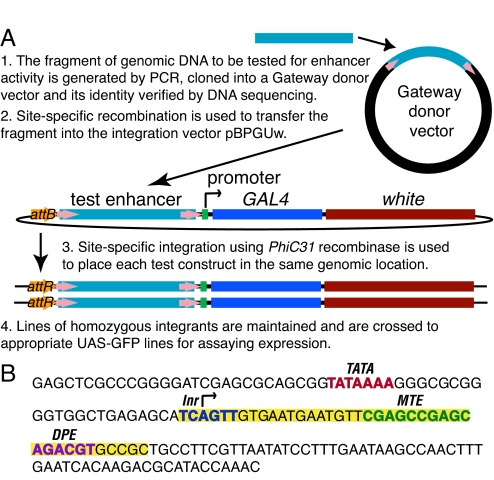
Fig. 1.
Strategy for constructing transgenic lines to test DNA segments for enhancer activity. (A) Diagram of the vectors and sequential cloning steps. (B) Sequence of the Drosophila synthetic core promoter (DSCP). Sequences highlighted in yellow were added to the promoter of the eve gene. The positions of known promoter motifs are indicated.
Enhancer activity could be tested by imaging the expression patterns that these fragments produce in transgenic animals. In the experiments described here, each enhancer drives the expression of the yeast transcription factor GAL4 (6, 7). We detected the expression of GAL4 either directly by whole mount in situ hybridization to its mRNA or by the ability of GAL4 protein to drive the expression of a UAS–GFP fusion gene whose products were then detected by immunocytochemistry and confocal microscopy of whole mount tissue (8).
Drosophila core promoters are ≈80 bp and contain the start site of transcription. An enhancer requires the presence of specific sequence motifs in the core promoter to function properly and the core promoters of different genes vary in their content of these motifs; thus, not all enhancers function efficiently with all core promoters (9). It has been shown that potent core promoters can be created by the incorporation of multiple core promoter motifs into a single promoter (10); more importantly, such promoters would be expected to respond to a wider range of enhancers than naturally occurring core promoters. To assay the enhancer elements from many different genes by using a single core promoter, we constructed a Drosophila synthetic core promoter (DSCP) that contains the TATA, Inr, MTE, and DPE sequence motifs (Fig. 1B).
We used the phiC31 site-specific integration system (11) to insert our constructs in the same orientation at the same genomic location. We selected an integration site, attP2 (Fig. S1) (11), which allows high levels of expression but does not appear to strongly influence the observed pattern of expression of the inserted construct; when inserted at this site, constructs that carry the DSCP but no enhancer lack detectable adult CNS expression (data not shown), many DNA fragments fail to drive any CNS expression, and there are no common pattern elements shared across large numbers of lines that do show CNS expression. Because of the consistent nature of the integration site, we could reliably compare the patterns of expression generated by different enhancer sequences and, once the expression pattern was determined, have confidence that we could drive the expression of other reporter genes in that pattern. Finally, having all constructs inserted at the same genomic location greatly simplifies subsequent genetic manipulations.
Evaluation of a Drosophila Synthetic Core Promoter.
We compared the expression patterns driven by 40 fragments derived from the dachshund (dac), earmuff (CG31670), and twin of eyeless (toy) genes when the fragments were paired either with their cognate promoter or with the DSCP (Tables S1–S3); the genomic extents of these fragments are shown as blue bars in Figs. 2A, 3A, and 4A. The dac, earmuff, and toy genes encode evolutionarily conserved transcription factors, with their vertebrate homologs being Dach, Fezl, and Pax6, respectively. An important feature of these genes for our purposes was that they were annotated as having unique transcription start sites, allowing us to select a single endogenous promoter for each.
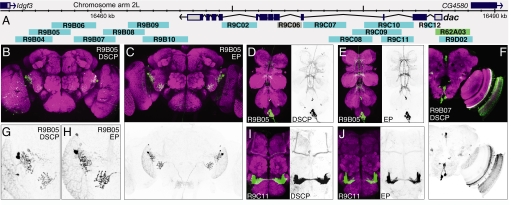
Fig. 2.
Testing fragments from the dac genetic region for enhancer activity with both the DSCP and the endogenous dac promoter. (A) Diagram of the chromosomal region surrounding the gene showing the structures of the transcription unit and those of adjacent genes (data taken from D. melanogaster genome sequence Release 4.1). The extent and position of the DNA segments that were tested for enhancer activity are shown as light blue bars below the map; R62A03 (green) is a promoter fusion, and we did not obtain data for R9C06 (gray). Total (B–E and G–J) or partial projections (F) of confocal images of the brain (B, C, F–J) or ventral nerve cord (D and E) of 2- to 5-day old adults of the indicated transgenic line. Fragments were tested with either the DSCP (B, D, F, G, and I) or the dac endogenous promoter (EP) (C, E, H, and J). In the color images, gene expression driven by the enhancer fragment is shown in green and the neuropil is counterstained in magenta (see SI Methods for details). Embryonic expression patterns are shown in Fig. S3. The gray scale images show only the enhancer-driven expression.
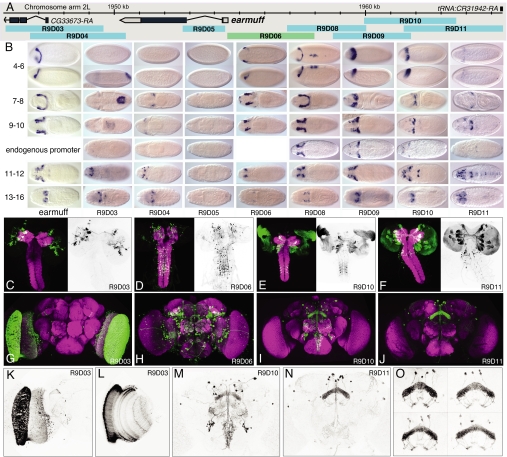
Fig. 3.
Patterns generated by fragments of the earmuff gene in embryos, larvae, and adults. (A) The genomic map of the earmuff locus. (B) Expression of the earmuff gene in embryonic stages 4–6, 7–8, 9–10, 11–12, and 13–16 visualized by whole mount in situ hybridization with a probe to earmuff mRNA shown adjacent to the expression produced by the fragments R9D03, R9D04, R9D05, R9D08, R9D09, R9D10, and R9D11 when placed in the enhancer test vector and fragment R9D06 as a promoter fusion. Transgene expression is visualized by whole mount in situ hybridization with a probe to GAL4 mRNA. The enhancer constructs shown use the DSCP; for stages 9–10, we also show data obtained with the endogenous earmuff promoter. Dorsal views are shown except for stages 4–6, where a lateral view is also shown below the dorsal view; anterior is at Left. (C, D, E, and F) Expression driven by the indicated fragment in late third instar larvae. The clusters of labeled cells seen in F represent distinct lineages of secondary neurons; this labeling is not maintained in the adult (J). (G–O) Expression in the adult brain of the indicated lines. A total projection (K) and single optical section (L) of the optic lobes of the brain shown in G. (O) Expression in the fan-shaped body of line R9D11 in four different brains.
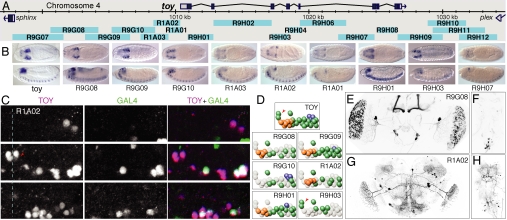
Fig. 4.
Patterns generated by fragments of the toy gene and a comparison with the expression pattern of the endogenous toy gene in the embryo. (A) Genomic map of the toy locus and the positions of the tested fragments. (B) Expression of the endogenous toy mRNA and the expression of GAL4 mRNA driven by the indicated nine fragments shown in stage 13–16 embryos; the other nine fragments shown in A did not drive detectable expression at this stage. Dorsal (Upper) and lateral (Lower) views are shown; anterior is at Left. (C) Endogenous toy protein (magenta in the merged image) and nuclear localized GFP (green in the merged image) expression driven by the R1A02 fragment in an abdominal CNS hemisegment of a stage 16 embryo (anterior, up; midline, dashed line). Three focal planes are shown: deep (Top), intermediate (Middle), and superficial (Bottom); see Fig. S2 for similar data on other fragments. (D) Diagrams of endogenous toy-positive neurons and the subset of toy-positive neurons in which each indicated fragment drives expression (deep neurons, blue; intermediate neurons, green; superficial neurons, orange); each fragment also drives expression in a reproducible set of toy-negative neurons that are not shown in these diagrams. The TM neurons are indicated by the red arrowheads. (E–H) Total projections of confocal images of the adult brain showing enhancer fragment driven expression in the brain (E and G) or ventral nerve cord (F and H) of lines R9G08 (E and F) and R1A02 (G and H).
To compare expression patterns, we established a controlled vocabulary for annotating patterns of axonal and dendritic projections. We first divided the brain into 45 identifiable brain structures, for example, antennal lobe, ellipsoid body, or great commissure. We separately scored each of these regions by using a zero to five scale for three parameters: intensity, distribution, and shape. (See SI Methods for a complete description of the controlled vocabulary.) Based on this scoring, we found that the variation between patterns generated with the two promoters was only slightly higher than that seen when comparing the same construct in multiple animals (see below); for 85% of the fragments, the patterns they drove when paired with the DSCP or their cognate promoter were identical in all three parameters in each brain structure. The patterns observed were larger or more pronounced in 10% of the cases with the DSCP and in 5% of the cases with the cognate promoter (see for example Fig. 2 B–E and G–J); however, even these differences were very subtle. In the embryo, we found that the DSCP routinely drove stronger expression (Fig. 3B). These results indicate that, for our purposes, the DSCP serves as an adequate surrogate for the core promoters of individual genes.
Relationship of the Expression Patterns Driven by Individual Fragments to the Expression Pattern of the Gene.
Our expectation from previous work was that individual fragments would drive subsets of the endogenous expression pattern of a gene (2–5). However, it was also likely that individual fragments, when taken out of context and freed from negatively acting elements, as well as from the necessity to compete with other enhancers for access to the core promoter, would drive expression in cells where the endogenous gene was not expressed. To address this possibility, we compared the embryonic expression patterns of earmuff (Fig. 3B) and toy (Fig. 4 B–D) with the patterns driven by individual fragments of these genes when combined with the DSCP. As expected, we found that individual fragments generally drove subsets of the wild-type expression pattern and, in sum, appeared to be able to reproduce all of the components of that pattern. However, with some of the fragments, we also saw reproducible expression in cells that do not express the endogenous gene.
The specificity and reproducibility of the patterns driven by individual fragments were illustrated by the expression patterns driven by six different fragments derived from the toy gene within the embryonic CNS (Fig. 4 C and D and Fig. S2). Endogenous toy protein was expressed in a highly stereotyped pattern of neurons within the stage 16 CNS: a pair of toy medial (TM) neurons, a superficial neuron cluster, a toy intermediate (TI) neuron cluster, and three toy deep lateral (TDL) neurons. Each fragment drove reproducible expression in a subset of the endogenous toy-positive neurons; for example, R9H03 was the only line expressed in the pair of TM neurons, whereas R9G10 was the only line expressed in all three TDL neurons. Each line also showed unique but reproducible expression in a subset of neurons that do not express toy protein; for example, R9G09 and R9G10 were the only toy-derived lines expressed in the RP2 motor neuron, whereas R9G09 and R9H01 were the only lines expressed in the U motor neurons (data not shown). We conclude that each of these fragments contains sequences that drive expression in a different, reproducible subset of the native toy pattern; in addition, when taken out of context, they also drive expression within distinct, reproducible subsets of neurons that do not normally express toy.
Enhancers Are Numerous with Each Controlling a Limited Subset of the Total Expression Pattern.
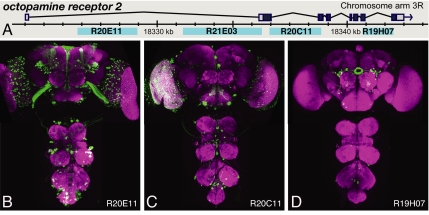
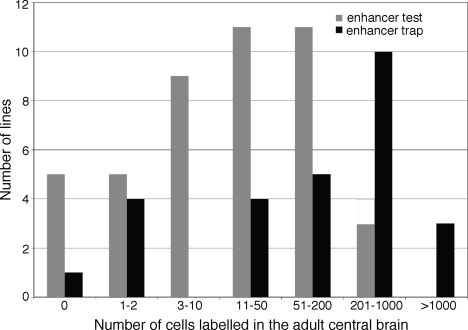
We tested 44 fragments derived from genes encoding the transcription factors Earmuff, TOY, DAC, and the G protein-coupled receptor octopamine receptor 2 (Fig. 5 and Table S4) for enhancer activity with the DSCP. Nearly 80% of these fragments generated expression patterns comprising 3 to 1,000 cells in the adult central brain; the central brain corresponds to the brain minus the optic lobes. The mean number of cells showing detectable expression was 95 in these lines; the median number of expressing cells was only 19. These cell numbers were much smaller than observed in a random sample of 27 enhancer trap lines where the observed mean and median were 370 and 180, respectively (Fig. 6); this sample was consistent with the expression patterns generally seen with enhancer trap lines (8). In enhancer trap lines, a transposon carrying a core promoter and a reporter gene is inserted randomly in the genome; the broader expression observed is likely a consequence of individual enhancer trap lines reporting the influence of multiple enhancers.
Fig. 5.
Distinct expression patterns generated by fragments of the octopamine receptor 2 gene. (A) Diagram of the genomic locus. (B–D) Expression driven by the indicated fragments in the adult brain and ventral nerve cord.
Fig. 6.
The patterns driven by individual fragments generally contain fewer cells than those found in enhancer trap lines. The gray bars in the histogram show the number of cells found in the patterns within the central brain of the adult generated by the 44 enhancer fragments shown in blue in Figs. 2A (dac), 3A (earmuff), 4A (toy), and 5A (octopamine receptor 2) genes. The black bars show the number of cells found in 27 enhancer trap lines that were chosen randomly from the unpublished collection of J. Simpson and B. Ganetsky and are typical of the type of patterns seen in other enhancer trap collections (8).
The patterns driven by a particular fragment are highly dynamic during development. For example, compare fragment R9D11 in the late larva (Fig. 3F) and the adult (Fig. 3J). The larva showed strong expression in ≈5% of the secondary lineages that produce the cells of the adult central brain, but in the adult central brain, expression is limited to approximately a dozen cells.
Further subdivision of the fragments will be required to determine the extent to which distinct enhancer activities within each fragment can be separated; in the ideal set of lines, each line would represent the expression pattern of a single enhancer. Overlapping fragments often showed overlapping patterns, suggesting that further subdivision would be possible. For example, compare the patterns driven by the fragments R9G08, R9G09, and R9G10, which drove expression in the TI cluster of embryonic TOY-expressing neurons; R9G08 and R9G10 drove expression in distinct subsets, whereas R9G09 drove expression in most or all of the TI neurons (see Fig. 4D and Fig. S2).
The Patterns Generated by the Same Enhancer in the Adult Brains of Different Animals Are Highly Reproducible.
If the GAL4-expressing lines we created were to have maximum utility, the patterns they produced would have to be highly reproducible from animal to animal. Variability might result from stochastic variation in gene expression activation or in anatomical variation. The degree of variability of adult brain anatomy between individual adult flies of the same genetic makeup has not been well documented. We compared the patterns generated by individual fragments in multiple individuals to address this question. By using the scoring scheme described above and blind study conditions, 95% of the isogenic brains from different animals were scored with identical annotations; even in the 5% that were not scored identically, the differences were subtle. This suggests that variation among animals at the granularity that we were scoring was minimal; however, we did observe that the positions of cell bodies vary much more than arborization patterns. This is illustrated in Fig. 3O, which shows the expression pattern of line R9D11 in the fan-shaped body of four animals.
Concluding Remarks.
Our results indicate that it should be possible to establish a collection of transgenic Drosophila lines, each directing expression to a small subset of cells in the adult brain, and that, in sum, would cover all cells in the brain. More than 80% of the 44 fragments we tested from four genes gave expression in the adult brain, suggesting that we would generate >4,000 lines with patterns from the analysis of our initial 5,200 putative enhancers. More than half of the fragments we tested gave expression in 10 to 200 cells: 0.03–0.7% of the 30,000 cells estimated to comprise the central brain (W. Pereanu, personal communication). We believe this fraction of cells is a useful number for anatomical, physiological, and behavioral studies. We recognize that the preferred cell number for behavioral studies is not known and will certainly depend on the particular behavior and assay; however, we expect that our lines, singly or in combination, will provide the versatility needed to generate expression patterns of the desired sizes. It is possible that the 2,000 lines we expect to generate having expression patterns within the 10 to 200 range of cell numbers will be adequate to represent all cells in the central brain in a variety of overlapping patterns. If not, it is straightforward to generate additional lines.
Our results reveal several features of genome organization. The number of distinct patterns we observed implies that there are likely to be >50,000 transcriptional enhancers in the Drosophila genome; >80% of the fragments we tested showed enhancer activity, it would take ≈50,000 such fragments to cover the entire genome, and many of these fragments are likely to carry multiple enhancers. A gene that is expressed in many tissues and developmental stages might have a single enhancer that controls all aspects of expression in a given stage or tissue; for example, an “adult brain enhancer”. Alternatively, the expression pattern at each stage and tissue might be generated by the sum of the actions of multiple enhancers, each controlling a subset of the pattern. Our data strongly favor the latter possibility. Furthermore, our results suggest that enhancers are reused throughout development, although the resolution of our current experiments was not sufficient to distinguish two closely linked enhancers from a single enhancer. It is also clear that enhancers, taken out of context, in addition to driving a subset of the expression pattern of the endogenous gene, often show highly stereotyped expression that is not displayed by the endogenous gene; this expression may reflect either the absence of competition between enhancers or the separation from repressive elements.
We believe that the lines we have generated, where a molecularly defined DNA fragment drives expression, have several advantages over existing tools for neuroanatomy and neurogenetics. The patterns we observed were less broad than those observed in enhancer trap lines, and our constructs were all at the same genomic location, facilitating subsequent genetic manipulations. It is also straightforward to attempt to produce smaller patterns by subdividing the fragments; these fragments were each large enough to carry several distinct enhancers. Most importantly, because our constructs were all inserted at precisely the same genomic location, the effects of local genomic environment on expression could be held constant. Thus, we should be able to change the gene whose expression is driven by a particular enhancer and have confidence that the expression pattern would remain the same. In this way, we could readily produce lines that drive, instead of GAL4, the expression of other transcription factors such as LexA (12), inhibitors of transcription factor function such as GAL80 (13), or recombinases such as Flp (7, 14) in the same patterns that we have determined for GAL4.
The small number of cells labeled in our lines can facilitate anatomical analysis, and the imaging of the expression patterns of several thousand such lines will provide a good overall view of the morphological range of neurons present in the fly brain and their projection patterns. Such studies can be aided by expression of markers directed to axons, dendrites, or synapses (15, 16). Stochastic labeling of individual cells within these patterns can be accomplished by using recombinases to facilitate detailed anatomy of individual cells (13, 14). It will be informative to examine the extent to which the cells that express a given enhancer share developmental, physiological, or functional properties and how shared activation of a given enhancer is related to the concept of cell type.
Finally, these lines will provide the ability to express genetically encoded indicators of function (17) or modifiers of neuronal activity (18) in well defined small subsets of neurons. We are optimistic that learning how many different behaviors are modified when the function of each of these small cell populations is altered will provide useful insights into the organization of neuronal circuits and information flow within the fly brain.
Methods
Standard molecular and histochemical methods were used; details of the constructs and protocols are given in SI Methods. Vectors are available from Addgene.
Supplementary Material
Acknowledgments.
Garson Tsang, Gina Dailey, Martha Evans-Holmes, and Soo Park provided technical assistance in the molecular biological aspects of this work; Richard Weiszmann and Amy Beaton assisted with in situ hybridization and imaging of embryos; Don Hall, Karen Hibbard, Amanda Cavallaro, Megan Hong, and Monti Mercer assisted with genetics and stock maintenance; and Rodney Simmons (Janelia Farm Research Campus, Howard Hughes Medical Institute, Ashburn, VA) provided media and general laboratory support. Eric Trautman (Janelia Farm Research Campus, Howard Hughes Medical Institute, Ashburn, VA) provided additional informatics tools. Phuong Chung and Julie Simpson taught us brain dissection and histochemical methods; Adina Bailey and Suzanna Lewis provided advice during the early stages of this work; and Susan Zusman and Michael Tworoger of Genetic Services, Inc., generated most of the transgenic lines. We thank Michael Layden for help in generating the data shown in Fig. 4C and Fig. S2. This work was supported by the Howard Hughes Medical Institute and by National Institutes of Health Grants GM076655–01A1 (to S.E.C.) and GM041249 (to J.T.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803697105/DCSupplemental.
References
- 1.Baker BS, Taylor BJ, Hall JC. Are complex behaviors specified by dedicated regulatory genes? Reasoning from Drosophila. Cell. 2001;105:13–24. doi: 10.1016/s0092-8674(01)00293-8. [DOI] [PubMed] [Google Scholar]
- 2.Hiromi Y, Kuroiwa A, Gehring WJ. Control elements of the Drosophila segmentation gene fushi tarazu. Cell. 1985;43:603–613. doi: 10.1016/0092-8674(85)90232-6. [DOI] [PubMed] [Google Scholar]
- 3.Harding K, Hoey T, Warrior R, Levine M. Autoregulatory and gap gene response elements of the even-skipped promoter of Drosophila. EMBO J. 1989;8:1205–1212. doi: 10.1002/j.1460-2075.1989.tb03493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goto T, Macdonald P, Maniatis T. Early and late periodic patterns of even skipped expression are controlled by distinct regulatory elements that respond to different spatial cues. Cell. 1989;57:413–422. doi: 10.1016/0092-8674(89)90916-1. [DOI] [PubMed] [Google Scholar]
- 5.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 6.Brand AH, Dormand EL. The GAL4 system as a tool for unraveling the mysteries of the Drosophila nervous system. Curr Opin Neurobiol. 1995;5:572–578. doi: 10.1016/0959-4388(95)80061-1. [DOI] [PubMed] [Google Scholar]
- 7.Duffy JB. GAL4 system in Drosophila: A fly geneticist's Swiss army knife. Genesis. 2002;34:1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- 8.Ito K, Okada R, Tanaka NK, Awasaki T. Cautionary observations on preparing and interpreting brain images using molecular biology-based staining techniques. Microsc Res Tech. 2003;62:170–186. doi: 10.1002/jemt.10369. [DOI] [PubMed] [Google Scholar]
- 9.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 10.Juven-Gershon T, Cheng S, Kadonaga JT. Rational design of a super core promoter that enhances gene expression. Nat Methods. 2006;3:917–922. doi: 10.1038/nmeth937. [DOI] [PubMed] [Google Scholar]
- 11.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci. 2006;9:703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- 13.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 14.Struhl G, Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- 15.Rolls MM, et al. Polarity and intracellular compartmentalization of Drosophila neurons. Neural Develop. 2007;2:7. doi: 10.1186/1749-8104-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, et al. Transmembrane/juxtamembrane domain-dependent Dscam distribution and function during mushroom body neuronal morphogenesis. Neuron. 2004;43:663–672. doi: 10.1016/j.neuron.2004.06.033. [DOI] [PubMed] [Google Scholar]
- 17.Reiff DF, et al. In vivo performance of genetically encoded indicators of neural activity in flies. J Neurosci. 2005;25:4766–4778. doi: 10.1523/JNEUROSCI.4900-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitamoto T. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J Neurobiol. 2001;47:81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.