Abstract
Psoriasis is a common inflammatory skin disease with a strong genetic component. We have analysed the genomic copy number polymorphism of the beta-defensin region on human chromosome 8 in 179 Dutch psoriasis patients and 272 controls, and in 319 German psoriasis patients and 305 controls. Comparisons in both cohorts show a significant association between higher genomic copy number for beta-defensin genes and the risk of psoriasis.
In humans, beta-defensins are small, secreted, antimicrobial peptides, which are encoded by DEFB genes in three main gene clusters: two on chromosome 20 and one on 8p23.11. Of the eight beta-defensin genes at 8p23.1, DEFB1 (encoding the protein hBD-1) and DEFB103 (encoding the protein hBD-3) are expressed constitutively in skin and DEFB4 (encoding the protein hBD-2) can be induced in cultured keratinocytes by cytokines or bacterial lipopolysaccharides2. The beta-defensin genes on 8p23.1, with the exception of DEFB1, but including DEFB4, SPAG11, DEFB103, DEFB104, DEFB105, DEFB106 and DEFB107, are on a large repeat unit (Supplementary Figure 1) that is variable in copy number3. Individuals have between 2 and 12 copies per diploid genome, with a modal copy number of four in the UK. The antimicrobial and pro-inflammatory nature of these beta-defensins suggests that quantitative variation in gene dosage might contribute to susceptibility to infectious and inflammatory disease4.
Psoriasis is a common inflammatory skin disease with a prevalence of about 2% in the populations of developed countries. It has both environmental and genetic parts to its etiology, and linkage analysis has been used to identify multiple loci and alleles that confer risk of the disease, with the strongest genetic effect at 6p21.3, where haplotypes carrying the HLA-Cw6 allele are associated with an increase in risk5. Psoriasis is characterized by red scaling elevated plaques commonly on elbows, knees and trunk. Histological examination of psoriatic lesions shows inflammation and disturbed epidermal differentiation. hBD-2 is induced as a part of the inflammatory response in skin, which, in psoriasis, is part of the regenerative maturation process involving hyperproliferation and induction of marker genes such as elafin and the cytokeratins 6, 16 and 17. In addition to their antimicrobial activity it has been shown that hBD-2 and other skin beta-defensins have cytokine-like properties6. The central role of these proteins in the innate immune system of the skin suggested that beta-defensin genes could be candidate genes for psoriasis susceptibility.
Copy number variation at the 8p23.1 beta-defensin cluster, commonly over the 2-7 copy range, poses greater challenges for accurate genotyping than at lower copy numbers. To investigate the relationship between beta-defensin copy number and susceptibility to psoriasis, we initially used a multiplex amplifiable probe hybridisation assay (MAPH, Supplementary Table 1) to determine copy number of the beta-defensin repeat per diploid genome3,7,8. Comparison of (unrounded) MAPH data from 190 Dutch cases and 303 controls suggested an association between increased defensin copy number and psoriasis (p = 1.65× 10−6, t-test). As an alternative assay for beta-defensin copy number we used the higher-throughput Paralogue Ratio Test (PRT) on the same set of samples; PRT typing of DEFB4 copy number uses specific co-amplification of a heat-shock protein pseudogene upstream of DEFB4 together with a single-copy paralogue on chromosome 5, and has been described previously9. Comparison of PRT results from 179 Dutch cases and 272 controls also suggested an association with psoriasis, but at a lower significance (p = 0.01, t-test).
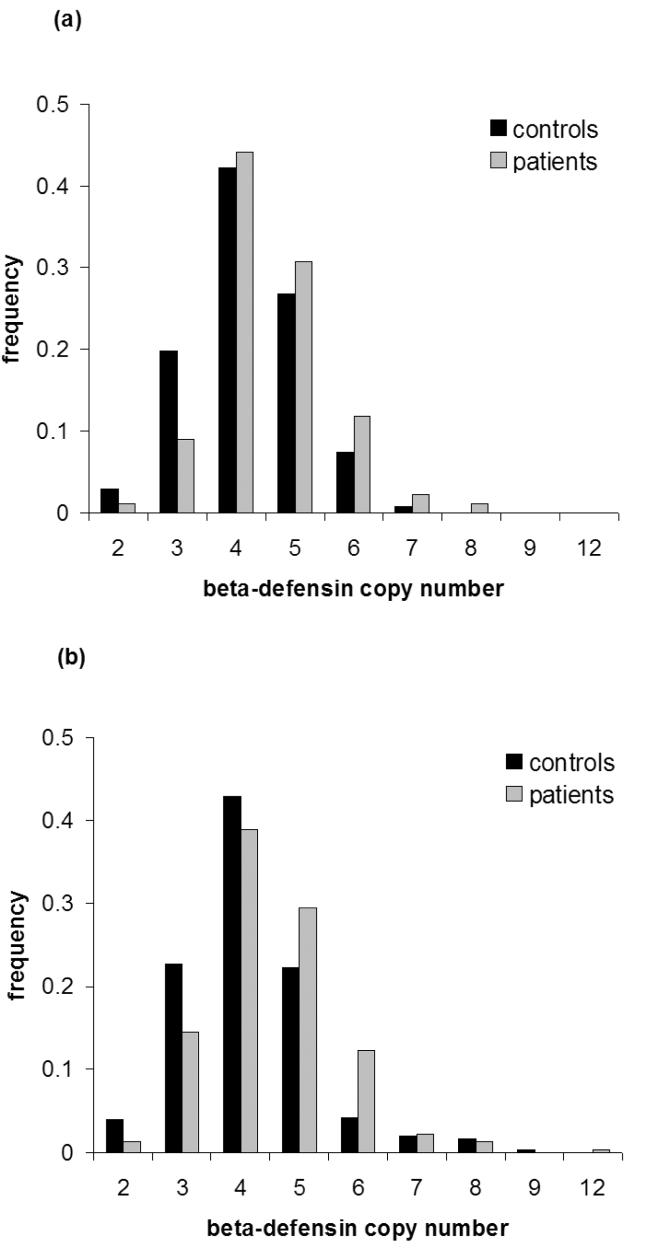
Although capable of high throughput, a single PRT assay has an error rate9 for DEFB4 copy number estimated at about 8%. True copy number values are likely to be integers. We therefore sought to improve the overall accuracy of copy number determination in this cohort by combining information into a consensus integer copy number for 179 cases and 272 controls, using data from MAPH, PRT, and ratios of multisite variants (MSVs)7,10 mapping around the DEFB4 gene (dbSNP reference numbers rs2740091, rs2737532 and rs3762040) (“REDVR”, see Supplementary Methods). Integer copy number agreed between MAPH/REDVR and first-pass PRT for 78% of samples, and for a further 11% on repeat PRT typing of discordant samples. This consensus integer copy number was significantly higher among cases than controls (Figure 1a, p = 7.8 × 10−5, t-test).
Figure 1.
Frequency distributions of beta-defensin genomic copy number.
a) Dutch controls and Dutch psoriasis patients.
b) German controls and German psoriasis patients.
As an independent test of this association we typed 305 controls and 319 patients from Germany using PRT alone. Analysis of PRT results showed a significant association with both unrounded (p = 9.02 × 10−6, t-test) and integer-rounded analyses (Figure 1b, p = 2.95 × 10−5, t-test). We do not believe that our findings can be attributed to differential genotyping bias11; in addition to the independent replication, important details of genotyping, including sample preparation, missing data and integer clustering are closely matched between cases and controls for both cohorts (see Supplementary Methods, sections C1 and C2). We have also made detailed comparisons between independent typing platforms for the Dutch samples, and believe that our copy number typing attains a high standard of accuracy (see Supplementary Methods, section C3). The full set of data for all samples typed is available as Supplementary Table 2. There was no significant difference between the Dutch and German cohorts for controls (p=0.165, t-test) or patients (p=0.95, t-test). Assuming a population prevalence of 2%, the relative risk of psoriasis for each copy number class can be inferred, with 1.00 representing the mean population risk (Table 1). Combining both cohorts, there is significant support (p=0.005, linear regression ANOVA, weighted by the square root of sample size) for a specifically linear model where each additional copy above two copies increases the relative risk, and the linear regression equation suggests that each copy adds 34 percentage points (95% CI 25%-43%) to the relative risk.
Table 1.
Influence of beta-defensin copy number on relative risk of psoriasis.
Counts are given in the table, together with the relative risk of psoriasis given the observed copy number, with a relative risk of 1.00 representing the population mean risk (2%).
| Copy number |
Dutch Controls |
Dutch Patients |
German Controls |
German Patients |
Combined Controls |
Combined Patients |
Copy number |
Population Frequency % |
Relative Risk* |
95% CI Relative Risk |
|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 8 | 2 | 12 | 4 | 20 | 6 | 2 | 3.5 | 0.31 | 0.12-0.77 |
| 3 | 54 | 16 | 69 | 46 | 123 | 62 | 3 | 21.3 | 0.51 | 0.37-0.71 |
| 4 | 115 | 79 | 131 | 124 | 246 | 203 | 4 | 42.6 | 0.84 | 0.67-1.05 |
| 5 | 73 | 55 | 68 | 94 | 141 | 149 | 5 | 24.4 | 1.08 | 0.83-1.40 |
| 6 | 20 | 21 | 13 | 39 | 33 | 60 | 6 or more | 8.1 | 1.69 | 1.16-2.48 |
| 7 | 2 | 4 | 6 | 7 | 8 | 11 | ||||
| 8 | 0 | 2 | 5 | 4 | 5 | 6 | ||||
| 9 | 0 | 0 | 1 | 0 | 1 | 0 | ||||
| 12 | 0 | 0 | 0 | 1 | 0 | 1 | ||||
| sum | 272 | 179 | 305 | 319 | 577 | 498 |
Relative risk values were calculated after pooling copy numbers of 6 or more into a single category. Bold type indicates a two-tailed significant difference from a relative risk of 1.00 at the 5% level. Population frequency shows an estimate of diploid copy number frequencies in native Northern Europeans deduced from the “combined controls” column.
We cannot exclude the possibility that the nucleotide state of an MSV actually causes susceptibility to psoriasis, and that copy number is only indirectly associated, as a proxy for this sequence variant. For the variants we have studied, in an extended Dutch cohort, psoriasis shows no association with rs3762040 (p=0.958) and only weak association (p=0.043) with rs2740091. However, we note that more than 200 substitutional variants are reported in dbSNP for this region and were not tested here.
Because of the cytokine-like properties of beta-defensins, either high resting levels or high induced levels may be a precipitating factor after minor skin injury, infection or some other environmental trigger. This could lead to an inappropriate inflammatory response generating the clinical symptoms typical of psoriasis. It has been shown that the level of hBD-2 in keratinocytes after induction by cytokines is correlated with its basal expression level12, and hBD-2, hBD-3 and hBD-4 have all been found to stimulate keratinocytes to release IL-8, IL-18 and IL-20, which are all proinflammatory cytokines that have an established role in the etiology of psoriasis13. The genes encoding hBD-2, hBD-3 and hBD-4 (DEFB4, DEFB103 and DEFB104 respectively) are all on the beta-defensin repeat and show the same copy number, so we cannot distinguish whether one gene or a combination of all three genes is responsible for the gene dosage association with psoriasis.
We have identified a psoriasis susceptibility locus not previously identified by linkage analysis. We therefore considered whether the strength of effect observed in our data should give rise to a detectable linkage signal. We simulated pedigrees with the same beta-defensin haplotype population frequencies and subject to the same dependence of risk on beta-defensin copy number genotype (Table 1, and Supplementary Methods). Only a minority of simulated studies involving 500 affected sib pairs detect linkage at even “suggestive” levels of significance (p<0.05). Similarly, simulation of sibling recurrence risk (λs), suggested that the beta-defensin locus accounts for a locus-specific λs of about 1.08. This low value largely reflects the common occurrence of the variation, and the fact that within pedigrees, total diploid copy number may not have a simple relationship with inheritance of parental alleles. At this level of effect, loci are undetectable by even large linkage studies, and may only be discovered by candidate gene association studies.
Copy number polymorphism of the cytokine CCL3L1 is reflected in expression levels and has been shown to influence susceptibility to HIV-1 infection14, and low copy number of complement C4 genes was associated with SLE in a recently published study15. We adopted PRT as a high-throughput method to genotype loci like these, with copy numbers of 2-12, to determine discrete copy numbers while maintaining an accuracy comparable to MAPH. Given the number of loci that vary in copy number across the genome, and the large number of these loci that are candidates for susceptibility to different diseases, large-scale copy number typing of case-control cohorts will be a priority.
Supplementary Material
Acknowledgements
We would like to thank Jess Tyson for help and advice, and Jane Hewitt and John Brookfield for comments on the manuscript. This work was funded by a Wellcome Trust Bioarchaeology Postdoctoral Fellowship awarded to EJH (grant 071024) and an NWO-Genomics grant to JS (050-10-102).
References
- 1.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003;3:710–20. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 2.Harder J, Bartels J, Christophers E, Schroder JM. A peptide antibiotic from human skin. Nature. 1997;387:861. doi: 10.1038/43088. [DOI] [PubMed] [Google Scholar]
- 3.Hollox EJ, Armour JAL, Barber JCK. Extensive normal copy number variation of a beta-defensin antimicrobial-gene cluster. Am. J. Hum. Genet. 2003;73:591–600. doi: 10.1086/378157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fellermann K, et al. A chromosome 8 gene-cluster polymorphism with low human beta-defensin 2 gene copy number predisposes to Crohn disease of the colon. Am. J. Hum. Genet. 2006;79:439–448. doi: 10.1086/505915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowcock AM, Krueger JG. Getting under the skin: the immunogenetics of psoriasis. Nat. Rev. Immunol. 2005;5:699–711. doi: 10.1038/nri1689. [DOI] [PubMed] [Google Scholar]
- 6.Niyonsaba F, Ogawa H, Nagaoka I. Human beta-defensin-2 functions as a chemotactic agent for tumour necrosis factor-alpha-treated human neutrophils. Immunology. 2004;111:273–81. doi: 10.1111/j.0019-2805.2004.01816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aldred PMR, Hollox EJ, Armour JAL. Copy number polymorphism and expression level variation of the human alpha-defensin genes DEFA1 and DEFA3. Hum. Mol. Genet. 2005;14:2045–2052. doi: 10.1093/hmg/ddi209. [DOI] [PubMed] [Google Scholar]
- 8.Hollox EJ, et al. Beta-defensin genomic copy number is not a modifier locus for cystic fibrosis. J. Neg. Res. BioMed. 2005;4:9. doi: 10.1186/1477-5751-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Armour JAL, et al. Accurate, high-throughput typing of copy number variation using paralogue ratios from dispersed repeats. Nucleic Acids Res. 2007;35:e19. doi: 10.1093/nar/gkl1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fredman D, et al. Complex SNP-related sequence variation in segmental genome duplications. Nature Genet. 2004;36:861–6. doi: 10.1038/ng1401. [DOI] [PubMed] [Google Scholar]
- 11.Clayton DG, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nature Genet. 2005;37:1243–1246. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- 12.Joly S, Organ CC, Johnson GK, McCray PB, Jr., Guthmiller JM. Correlation between beta-defensin expression and induction profiles in gingival keratinocytes. Mol. Immunol. 2005;42:1073–84. doi: 10.1016/j.molimm.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Niyonsaba F, Ushio H, Nagaoka I, Okumura K, Ogawa H. The human beta-defensins (−1, −2, −3, −4) and cathelicidin LL-37 induce IL-18 secretion through p38 and ERK MAPK activation in primary human keratinocytes. J. Immunol. 2005;175:1776–84. doi: 10.4049/jimmunol.175.3.1776. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez E, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y, et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am. J. Hum. Genet. 2007;80:1037–1054. doi: 10.1086/518257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.