Abstract
Lamin A mutations cause many diseases, including cardiomyopathies and Progeria Syndrome. The covalent attachment of small ubiquitin-like modifier (SUMO) polypeptides regulates the function of many proteins. Until now, no examples of human disease-causing mutations that occur within a sumoylation consensus sequence and alter sumoylation were known. We show that lamin A is sumoylated at lysine 201 and that two lamin A mutants associated with familial dilated cardiomyopathy, E203G and E203K, exhibit decreased sumoylation. E203 occupies the conserved +2 position in the sumoylation consensus ΨKXE. Lamin A mutants E203G, E203K, and K201R all exhibit a similar aberrant subcellular localization and are associated with increased cell death. Fibroblasts from an individual with the E203K lamin A mutation also exhibit decreased lamin A sumoylation and increased cell death. These results suggest that SUMO modification is important for normal lamin A function and implicate an involvement for altered sumoylation in the E203G/E203K lamin A cardiomyopathies.
Introduction
The lamin A protein plays an important role in the structure and function of the nucleus, and mutations in the lamin A gene cause a large number of different human diseases, including cardiomyopathies, muscular dystrophies, and Hutchinson-Gilford Progeria Syndrome (Broers et al., 2006; Capell and Collins, 2006; Mattout et al., 2006; Parnaik and Manju, 2006). Covalent attachment of small ubiquitin-like modifier (SUMO) proteins to lysine residues in target proteins, or sumoylation, is an important regulator of protein functional properties (Hay, 2005; Bossis and Melchior, 2006; Kerscher et al., 2006). SUMO proteins are covalently attached to target lysine residues by the SUMO E2 enzyme ubc9, and these substrate lysines are typically found within the consensus sequence ΨKXE (Ψ represents hydrophobic amino acids; Desterro et al., 1997; Johnson and Blobel, 1997; Rodriguez et al., 2001; Sampson et al., 2001). Cells express three major SUMO paralogues, SUMO-1, SUMO-2, and SUMO-3, with SUMO-2 and -3 being much more similar to each other than to SUMO-1 (Hay, 2005; Kerscher et al., 2006; Bossis and Melchior, 2006).
Using a yeast two-hybrid screen, a previous study identified an interaction between lamin A and ubc9, the SUMO E2 protein (Zhong et al., 2005). Based on this interaction, we hypothesized that the lamin A protein could be a target of sumoylation. The purpose of the experiments in this present study was to determine whether lamin A is indeed sumoylated in cells and, if so, what role this modification plays in regulating the function of this lamin.
Results and discussion
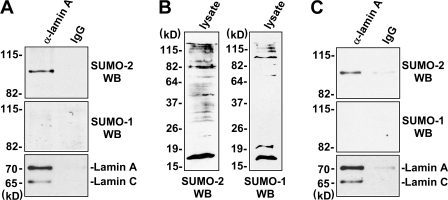
First, we sought to test for sumoylation of endogenous lamin A by performing immunoprecipitation of HeLa cell extracts using lamin A antibodies, followed by Western using antibodies against SUMO-1 or SUMO-2/SUMO-3 (because of the similarity of SUMO-2 and -3, it is likely that both of these SUMO proteins are recognized by this antibody). The results suggest that lamin A is SUMO modified and that it is preferentially modified by SUMO-2 compared with SUMO-1 (Fig. 1 A). The results in Fig. 1 C indicate that lamin A protein in extracts of mouse heart is also sumoylated and that, like lamin A that is present in HeLa cell extracts, SUMO-2 appears to be the predominant SUMO protein attached to this protein.
Figure 1.
Endogenous lamin A is sumoylated. (A) Extracts of HeLa cells were subjected to immunoprecipitation using anti–lamin A antibodies followed by Western blot assays using antibodies against SUMO-2, SUMO-1, or lamin A (different from those used for immunoprecipitation). (B) Western blot assays of HeLa cell lysates used for the immunoprecipitations using anti–SUMO-2 or –SUMO-1 antibodies. (C) Extracts prepared from mouse heart were subjected to immunoprecipitation using anti–lamin A antibodies followed by Western blot assays using antibodies against SUMO-2, SUMO-1, or lamin A (different from those used for immunoprecipitation).
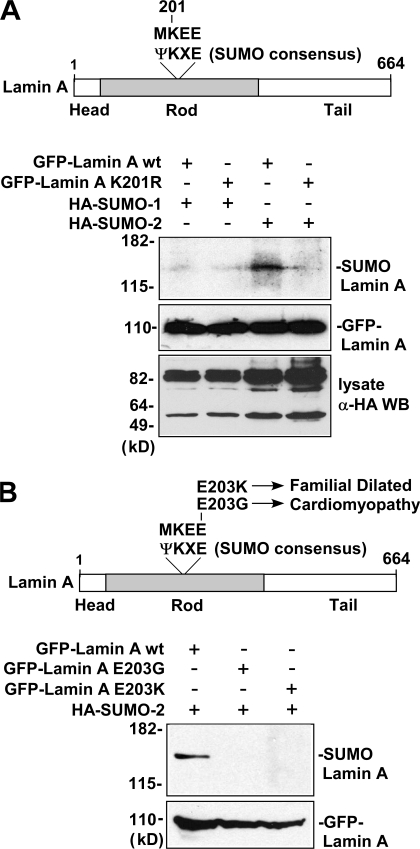
Analysis of the lamin A amino acid sequence revealed a match to the sumoylation consensus sequence ΨKXE (MKEE) surrounding lysine 201 in the rod-containing domain of lamin A (Fig. 2 A, top). To test whether sumoylation of the lamin A is occurring at lysine 201, HeLa cells were transfected with mammalian expression plasmids encoding GFP fusion constructs of wild-type lamin A and lamin A in which this lysine was changed to a nonsumoylatable arginine (K201R), along with expression constructs encoding HA-tagged SUMO-1 or -2. Extracts of the transfected cells were subjected to immunoprecipitation with anti-GFP antibodies followed by anti-HA Western blot. The results of this experiment, in agreement with the results obtained for endogenous lamin A, indicated that the wild-type lamin A protein is covalently modified by SUMO-2 but not as efficiently sumoylated by SUMO-1 (Fig. 2 A, bottom). The results also show that the modification by SUMO-2 is not observed for the K201R lamin A mutant protein, suggesting that lysine 201 is the site of SUMO-2 attachment in this protein.
Figure 2.
Lamin A is sumoylated at lysine 201 by SUMO-2, and E203G and E203K mutant lamin A proteins exhibit decreased sumoylation. (A) The top shows the location of a match (MKEE) to the sumoylation site consensus sequence (ΨKXE) surrounding lysine 201 in the rod-containing domain of lamin A. In the bottom, extracts of HeLa cells transfected with GFP fusion constructs of wild-type lamin A or K201R mutant lamin A and HA-tagged SUMO-1 or -2 plasmids were subjected to immunoprecipitation using anti-GFP antibodies followed by anti-HA Western blot. Western blots of the lysates using anti-GFP and anti-HA antibodies show levels of GFP–lamin A proteins and HA–SUMO-1/HA–SUMO-2 proteins (incorporated into major sumoylated proteins), respectively. (B) The top shows the location of the E203G and E203K mutations in lamin A associated with familial dilated cardiomyopathy at the conserved glutamic acid residue of the sumoylation site consensus sequence (ΨKXE) surrounding lysine 201. In the bottom, extracts of HeLa cells transfected with GFP fusion constructs of wild-type lamin A, E203G mutant lamin A, or E203K mutant lamin A along with HA-tagged SUMO-2 were subjected to immunoprecipitation with anti-GFP antibodies followed by anti-HA Western blot. Levels of transfected GFP–lamin A proteins in the cell lysates were determined by Western blot using anti-GFP antibody.
The glutamic acid residue at the fourth position in the sumoylation consensus sequence ΨKXE is known to be important for the efficiency of SUMO addition to the nearby lysine in this sequence (Rodriguez et al., 2001; Sampson et al., 2001). Relevant to this, two different disease-associated mutations of the human lamin A gene have been identified that change the glutamic acid at this position (E203) in the sumoylation consensus sequence of this protein to a different amino acid (Fig. 2 B, top; Fatkin et al., 1999; Jakobs et al., 2001). In one of these mutants, glutamic acid 203 is changed to glycine (E203G), whereas in the other, it is changed to lysine (E203K). Both the E203G and E203K mutations of lamin A are associated with familial dilated cardiomyopathy and conduction system disease (Fatkin et al., 1999; Jakobs et al., 2001), but the underlying molecular mechanism by which these mutations alter lamin A function is not known. Based on the results in the previous paragraph demonstrating that sumoylation of the lamin A protein occurs at lysine 201 and on previous results indicating the importance of glutamic acid for sumoylation at the preceding lysine of the ΨKXE sumoylation consensus sequence (Rodriguez et al., 2001; Sampson et al., 2001), we hypothesized that the E203G and E203K mutations could possibly mediate their deleterious effects by resulting in decreased sumoylation of the lamin A protein. To test the feasibility of this hypothesis, we performed a transfection immunoprecipitation experiment similar to that shown in Fig. 2 A, except that here the sumoylation of wild-type GFP–lamin A was compared with that of GFP–lamin A constructs containing the E203G or E203K mutations. The results of this experiment, shown in Fig. 2 B (bottom), indicate that both the E203G and E203K mutant lamin A proteins exhibit decreased sumoylation compared with the wild-type lamin A protein.
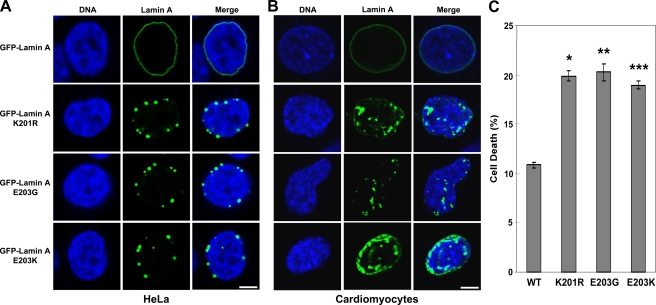
Sumoylation plays an important role in regulating the functional properties of target proteins in cells (Hay, 2005; Bossis and Melchior, 2006; Kerscher et al., 2006). The wild-type lamin A protein exhibits a characteristic pattern of localization at the nuclear periphery (Broers et al., 2006; Capell and Collins, 2006; Mattout et al., 2006; Parnaik and Manju, 2006). We hypothesized that sumoylation of lamin A at lysine 201 may be important for this localization pattern. To test this hypothesis, HeLa cells were transfected with the wild-type or K201R lamin A GFP fusion expression plasmids and then examined by fluorescence microscopy. As shown in Fig. 3 A, the wild-type lamin A GFP fusion protein exhibits the typical pattern of relatively continuous nuclear peripheral localization. However, the K201R lamin A GFP fusion protein shows an altered localization pattern, with the mutant protein appearing to concentrate into foci. These results suggest that sumoylation of lamin A is important for the normal pattern of subcellular localization of this protein. Analysis of the subcellular localization of GFP fusion constructs of the E203G or E203K mutant lamin A proteins by fluorescence microscopy reveals that both of these mutant proteins exhibit altered localization patterns similar to that of the K201R mutant lamin A, in which these proteins are concentrated in foci, in contrast to the more continuous appearance of the wild-type lamin A at the nuclear periphery (Fig. 3 A). The pattern we observed for the E203G lamin A is similar to that observed by a previous study (Lloyd et al., 2002). To our knowledge, the localization of the E203K lamin A has not previously been reported. The percentages of cells exhibiting abnormal localization of the wild-type and mutant GFP–lamin A proteins are shown in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200712124/DC1).
Figure 3.
SUMO site K201R mutant lamin A and cardiomyopathy-associated E203G and E203K mutant lamin A proteins exhibit similar patterns of aberrant localization. (A and B) Wild-type, K201R, E203G, and E203K lamin A GFP fusion expression plasmids were transfected into HeLa cells (A) or mouse embryonic cardiomyocytes (B), and the subcellular localization of the GFP–lamin A proteins was examined by confocal fluorescence microscopy. DNA was visualized by staining with HOECHST 33342. Bars, 5 μm. (C) Wild-type, K201R, E203G, or E203K lamin A GFP fusion expression plasmids were transfected into HeLa cells, which were then analyzed by trypan blue assay to measure cell death. Data are shown as means ± SEM. (*, P < 0.0001; **, P < 0.0002; ***, P < 0.0001 [for lamin A mutants vs. wild type]) and are each from three datasets.
To confirm the effects of altered localization of the cardiomyopathy-associated E203G and E203K lamin A mutants in a more physiologically relevant cell type, we repeated the experiments shown in Fig. 3 A in mouse embryonic cardiomyocytes. The results, shown in Fig. 3 B, indicate that the K201R, E203G, and E203K lamin A mutants all exhibit aberrant localization patterns in the cardiomyocytes (compared with wild-type lamin A) that are similar to those observed in HeLa cells. Percentages of cells exhibiting abnormal localization of wild-type and mutant GFP–lamin A are shown in Table S1.
In light of the defective sumoylation and aberrant localization patterns of the K201R, E203G, and E203K lamin A mutant proteins, we hypothesized that expression of these lamin A mutant proteins could result in decreased cell viability. The results shown in Fig. 3 C indicate that this appears to be the case, as all three of these mutant lamin A proteins are associated with increased cell death.
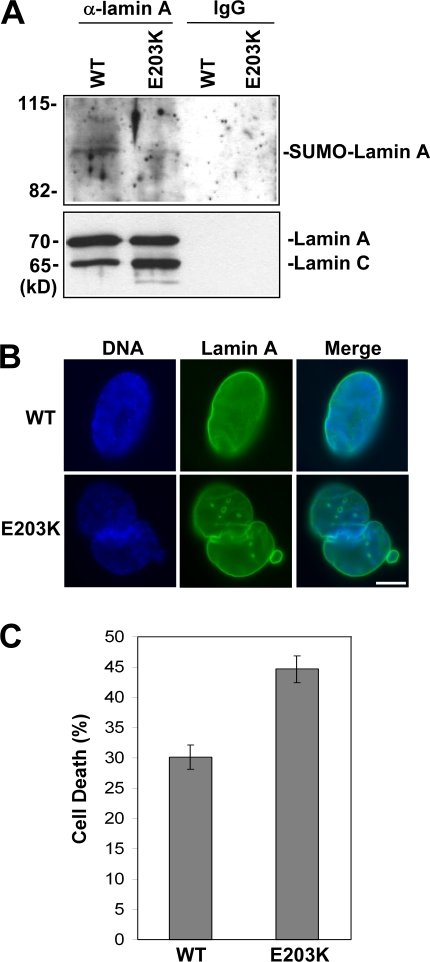
In the final set of experiments, we examined skin fibroblasts obtained from a patient that is heterozygous for the E203K lamin A mutation, which, as already described, is associated with cardiomyopathy (Jakobs et al., 2001). Immunoprecipitation of lamin A from these cells followed by SUMO-2 Western blot indicates that, as expected, levels of sumoylation of lamin A are reduced in cells from this individual compared with cells of a normal individual (Fig. 4 A). This individual is heterozygous for the E203K lamin A mutation, thus we expect that lamin A sumoylation was decreased but not eliminated. Further, immunofluorescence analysis of lamin A indicated more cells showing abnormal lamin A localization and nuclear morphology for the E203K fibroblasts compared with normal fibroblasts (Fig. 4 B). The percentages of normal and E203K fibroblasts exhibiting abnormal lamin A localization/nuclear morphology are shown in Table S1. The E203K fibroblasts also exhibit increased cell death compared with the control fibroblasts (Fig. 4 C).
Figure 4.
Fibroblasts from an individual with E203K lamin A mutation exhibit decreased lamin A sumoylation, altered lamin A localization/nuclear morphology, and increased cell death. (A) Extracts prepared from skin fibroblasts of a normal individual and individual heterozygous for the E203K lamin A mutation were subjected to immunoprecipitation using anti–lamin A antibodies followed by Western blot assay using anti–SUMO-2 antibodies or anti–lamin A antibodies (different from those used for immunoprecipitation). (B) Skin fibroblasts of a normal individual and the individual heterozygous for the E203K lamin A mutation were subjected to immunofluorescence analysis using anti–lamin A antibodies. DNA was visualized by staining with HOECHST 33342. Bar, 5 μm. (C) Skin fibroblasts of a normal individual and E203K lamin A individual were analyzed by trypan blue assay to measure cell death. Data are shown as means ± SEM (P < 0.0038 for E203K lamin A mutant cells vs. wild-type cells) and are from three datasets.
The results presented in this paper indicate that lysine 201 of lamin A is a target of covalent modification by the SUMO-2 protein and that this sumoylation is important for the normal pattern of subcellular localization of the lamin A protein. The results of these experiments also show that lamin A sumoylation is decreased in transfected mutant E203G and E203K lamin A proteins that cause familial dilated cardiomyopathies and in lamin A of fibroblasts of individuals with the E203K mutation. To our knowledge these are the first examples of human disease-causing mutations occurring in a crucial residue of a sumoylation consensus sequence and resulting in decreased sumoylation of the mutant protein.
The results also indicate that the mutant E203G and E203K lamin A proteins exhibit altered subcellular localization patterns that are very similar to that of the SUMO attachment site mutant K201R lamin A protein. These results suggest a role for sumoylation in the correct localization of lamin A in the cell. Further, cells transfected with the E203G and E203K lamin A proteins and skin fibroblasts of individuals with the E203K mutation exhibit higher levels of cell death compared with cells transfected with wild-type lamin A or normal skin fibroblasts, respectively. Together, these results suggest that sumoylation is important for normal lamin A function and support a role for altered sumoylation in the underlying molecular mechanism of familial dilated cardiomyopathies associated with the E203G/E203K lamin A mutations.
Materials and methods
Cell culture and plasmids
HeLa cells were cultured in DME medium (Mediatech, Inc.) supplemented with 10% FBS and 100× antibiotic-antimycotic (Invitrogen) in 5% CO2. Human skin fibroblasts (provided by R. Hershberger, University of Miami, Miami, FL) were cultured in the same media supplemented with 2 mM glutamine. Transfection of HeLa cells was performed using Effectene transfection reagent (QIAGEN), according to the manufacturer's protocol. Immunoprecipitation analysis and fluorescence microscopy were performed 48 h after the transfection. The GFP–lamin A plasmid was constructed from the pcDNA3.1-LMNA plasmid (gift of N. Zhong and W.T. Brown, Institute for Basic Research in Developmental Disabilities, Staten Island, NY). The coding region of the lamin A protein was cut out with EcoRI and BamHI digestion and ligated into pEGFP-C2 vector (Clontech Laboratories, Inc.). Mutagenesis PCR was performed to generate GFP–lamin A K201R, E203G, and E203K mutants. The primers used for the mutagenesis were as follows (only top primers are listed, bottom primers are the reverse complements of each of these): K201R, 5′-CTG CAG ACC ATG AGG GAG GAA CTG GAC-3′; E203G, 5′-ACC ATG AAG GAG GGA CTG GAC TTC CAG-3′; and E203K, 5′-ACC ATG AAG GAG AAA CTG GAC TTC CAG-3′. HA–SUMO-1 and –SUMO-2 were expressed using pcDNA3–HA–SUMO-1 and –HA–SUMO-2 plasmids (provided by K. Orth, University of Texas Southwestern, Dallas, TX).
Immunoprecipitation analysis
For immunoprecipitation experiments, the cell pellets of the transfected HeLa cells or the human skin fibroblasts were resuspended in 500 μl RIPA buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.4, 1% NP-40, 0.2% SDS, 0.25% sodium deoxycholate, and 1 mM EDTA) with protease inhibitor cocktail (Roche), 1 mM DTT, and 20 mM N-ethylmaleimide (added fresh). Cell lysis was performed by sonication, after which the sample was incubated on ice for 20 min. After centrifugation at 10,000 rpm at 4°C for 10 min, the supernatant was transferred to a fresh tube, and 20 μl of the whole cell lysate was removed for analysis of the level of transfected GFP–lamin A. 300 μl of 50% protein G–Sepharose slurry (GE Healthcare) was washed three times with PBS and resuspended in RIPA buffer to make a 50% slurry. The cell lysate was precleared by mixing it with 150 μl of this slurry and 5 μg of goat IgG and incubating at 4°C for 60 min. After centrifugation at 4,000 rpm at 4°C for 1 min, the supernatant was transferred to a fresh tube and mixed with 5 μg of anti-GFP antibody (Bethyl Laboratories, Inc.) or anti–lamin A antibody (BD Biosciences). After incubation at 4°C for 60 min, 150 μl of 50% protein G–Sepharose slurry was added and incubated at 4°C for 60 min. The beads were washed with RIPA buffer four times and then boiled in 50 μl of 4× SDS-PAGE loading buffer. After brief centrifugation, the supernatants were subjected to SDS-PAGE and Western blot using the antibodies described in the next section.
Western blot analysis and antibodies
SDS-PAGE and Western blot were performed according to standard procedures. The antibodies and dilutions used to probe the Western blots were as follows. Goat anti-GFP antibody (Bethyl Laboratories, Inc.) was used at 1:2,000, mouse anti-HA antibody (gift from D. Andres laboratory, University of Kentucky, Lexington, KY) was used at 1:2,000, goat anti–SUMO-1 antibody (Bethyl Laboratories, Inc.) was used at 1:1,000, rabbit anti–SUMO-2 antibody (Abgent) was used at 1:500, and rabbit anti–lamin A antibody (Abcam) was used at 1:500.
Fluorescence and immunofluorescence microscopy
Cells were seeded on coverslips. At 48 h after transfection with the wild-type or point mutant GFP–lamin A expression constructs, HOECHST 33342 and verapamil were added to the medium to final concentrations of 5 and 50 μg/ml, respectively. After incubation at 37°C for 30 min, the coverslips were washed twice with PBS and then incubated in 3.7% PFA at room temperature for 20 min. After two washes with PBS and a wash with distilled water, the coverslips were wicked on a kimwipe to partially dry and then mounted onto a slide spotted with 15 μl of VectorShield (Vector Laboratories). For immunostaining, after two washes with PBS, the coverslips were incubated in PBS containing 3% BSA at 37°C for 1 h and then incubated with rabbit anti–lamin A antibody (1:1,000) at 37°C for 30 min. After the coverslip was washed twice with PBS, it was incubated with Fluorescein anti–rabbit IgG (1:200) at 37°C for 30 min. After two washes with PBS and a brief wash with distilled water, the coverslip was mounted onto a slide spotted with 15 μl of VectorShield. Excess fluid was wicked from the coverslips, and the edges of the coverslip were sealed with fingernail polish. The fluorescence was then visualized using a confocal (TCS SP5; Leica) or fluorescent (Nikon) microscope with a Spotcam digital-imaging camera (Nikon). Figures were made from the images using Photoshop (Adobe).
Trypan blue cell viability assay
Cells were collected by centrifugation at 1,000 rpm for 10 min at 4°C, and the pellet was washed twice with PBS. The cell pellet was then resuspended in PBS to a concentration of ∼106 cells/ml. A one to one dilution of the suspension was prepared using a solution containing 0.4% trypan blue stain (Invitrogen), and the suspension was then loaded into the counting chamber of a hemacytometer. The number of stained cells, as well as the total number of cells, was counted, and the percentage of stained cells was taken to represent the percentage of cell death. Three datasets were analyzed for each experiment.
Statistical analysis
Statistical significance was determined using Student's t test. A p-value of <0.05 was considered to be statistically significant.
Preparation of cardiomyocytes
These cells were provided by W. Lester and J. Satin (University of Kentucky, Lexington, KY). All animals and animal procedures used in this study were approved by the University of Kentucky Institutional Animal Care and Use Committee. E16 mouse (ICR outbred strain; Harlan) hearts were dissected free of connective tissues, and ventricles were separated from conotruncus and sinus venosus or atria. Cells were enzymatically dispersed and cultured as previously described (Cribbs et al., 2001). In brief, 10–40 embryos were minced and quickly transferred to nominally Ca2+-free digestion buffer containing 0.5 mg/ml collagenase (type II; Worthington) and 1 mg/ml pancreatin for two 15-min cycles. Digested tissue yielded a large fraction of single spontaneously beating cells in culture media consisting of DME containing 10% FBS.
Online supplemental material
Table S1 contains quantitation analysis for the experiments in Fig. 3 (A and B) and Fig. 4 B. This table shows the percentages of HeLa cells or cardiomyocytes transfected with GFP lamin A wild-type, K201R, E203G, or E203K mutants that exhibit abnormal lamin A localization and the percentages of normal versus E203K lamin A fibroblasts showing abnormal lamin A localization/nuclear morphology. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200712124/DC1.
Supplementary Material
Acknowledgments
We are very grateful to Dr. Ray Hershberger for providing skin fibroblasts from normal and E203K lamin A individuals, Dr. Nanbert Zhong and Dr. W. Ted Brown for the pcDNA3.1-LMNA plasmid, Dr. Kim Orth for the HA–SUMO-1 and HA–SUMO-2 expression plasmids, Dr. Doug Andres for anti-HA antibodies, and William Lester and Dr. Jon Satin for the kind gift of embryonic cardiomyocytes. We also thank members of our laboratory for insightful discussions.
This research was supported by National Institutes of Health grant GM64606 to K.D Sarge.
Abbreviation used in this paper: SUMO, small ubiquitin-like modifier.
References
- Bossis, G., and F. Melchior. 2006. SUMO: regulating the regulator. Cell Div. 1:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broers, J.L., F.C. Ramaekers, G. Bonne, R.B. Yaou, and C.J. Hutchison. 2006. Nuclear lamins: laminopathies and their role in premature ageing. Physiol. Rev. 86:967–1008. [DOI] [PubMed] [Google Scholar]
- Capell, B.C., and F.S. Collins. 2006. Human laminopathies: nuclei gone genetically awry. Nat. Rev. Genet. 7:940–952. [DOI] [PubMed] [Google Scholar]
- Cribbs, L.L., B.L. Martin, E.A. Schroder, B.B. Keller, B.P. Delisle, and J. Satin. 2001. Identification of the t-type calcium channel CaV3.1d in developing mouse heart. Circ. Res. 88:403–407. [DOI] [PubMed] [Google Scholar]
- Desterro, J.M., J. Thomson, and R.T. Hay. 1997. Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 417:297–300. [DOI] [PubMed] [Google Scholar]
- Fatkin, D., C. MacRae, T. Sasaki, M.R. Wolff, M. Porcu, M. Frenneaux, J. Atherton, H.J. Vidaillet, S. Spudich, U. De Girolami, et al. 1999. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N. Engl. J. Med. 341:1715–1724. [DOI] [PubMed] [Google Scholar]
- Hay, R.T. 2005. SUMO: a history of modification. Mol. Cell. 18:1–12. [DOI] [PubMed] [Google Scholar]
- Jakobs, P.M., E.L. Hanson, K.A. Crispell, W. Toy, H. Keegan, K. Schilling, T.B. Icenogle, M. Litt, and R.E. Hershberger. 2001. Novel lamin A/C mutations in two families with dilated cardiomyopathy and conduction system disease. J. Card. Fail. 7:249–256. [DOI] [PubMed] [Google Scholar]
- Johnson, E.S., and G. Blobel. 1997. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 272:26799–26802. [DOI] [PubMed] [Google Scholar]
- Kerscher, O., R. Felberbaum, and M. Hochstrasser. 2006. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22:159–180. [DOI] [PubMed] [Google Scholar]
- Lloyd, D.J., R.C. Trembath, and S. Shackleton. 2002. A novel interaction between lamin A and SREBP1: implications for partial lipodystrophy and other laminopathies. Hum. Mol. Genet. 11:769–777. [DOI] [PubMed] [Google Scholar]
- Mattout, A., T. Dechat, S.A. Adam, R.D. Goldman, and Y. Gruenbaum. 2006. Nuclear lamins, diseases and aging. Curr. Opin. Cell Biol. 18:335–341. [DOI] [PubMed] [Google Scholar]
- Parnaik, V.K., and K. Manju. 2006. Laminopathies: multiple disorders arising from defects in nuclear architecture. J. Biosci. 31:405–421. [DOI] [PubMed] [Google Scholar]
- Rodriguez, M.S., C. Dargemont, and R.T. Hay. 2001. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276:12654–12659. [DOI] [PubMed] [Google Scholar]
- Sampson, D.A., M. Wang, and M.J. Matunis. 2001. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276:21664–21669. [DOI] [PubMed] [Google Scholar]
- Zhong, N., G. Radu, W. Ju, and W.T. Brown. 2005. Novel progerin-interactive partner proteins hnRNP E1, EGF, Mel 18, and UBC9 interact with lamin A/C. Biochem. Biophys. Res. Commun. 338:855–861. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.