Abstract
Nucleophosmin (NPM) is frequently mutated in acute myeloid leukemias and is thought to act as both a proto-oncogene and a tumor suppressor. Although genetic and molecular evidence has shed light on the mechanisms of NPM-mediated tumor suppression, the potential role of NPM mutants as oncogenes remains ill defined. Now, new data provide a straightforward mechanism for this latter function, as NPM is shown to regulate the stability and the function of MYC. Remarkably, the same leitmotif of “placing a critical cell regulator in the wrong place at the wrong time” appears to underscore all the cancer-promoting activities of mutated NPM.
Nucleophosmin (NPM; also known as B23, NO38, or numatrin) is a nucleolar protein that shuttles continuously between the nucleus and the cytoplasm (for review see Grisendi et al., 2006). Because of its nucleolar localization, intrinsic RNase activity, and association with maturing preribosomal ribonucleoproteins, NPM has been proposed to regulate ribosomal RNA transcription/processing and the transport of preribosomal particles to the cytoplasm (Grisendi et al., 2006). One major feature of NPM is to display molecular chaperone activity: it functions as a histone chaperone, favoring DNA-histone and nucleosome assembly in vitro, and also interacts with a wide range of unfolded proteins, inducing proper folding in the active state (Grisendi et al., 2006). In vivo, NPM interacts with many growth regulators, including the tumor suppressors p53 and ARF, and the HDM2 (Mdm2 in mouse) oncogene. Not surprisingly, this translates into pleiotropic biological effects, including regulation of nuclear export, DNA transcription, DNA repair, and cell proliferation and survival (Grisendi et al., 2006).
Much of the interest in NPM derives from its involvement in cancer. NPM is frequently overexpressed in carcinomas and is involved in chromosome translocations in hematological malignancies, where it forms fusion proteins with different partners (for review see Grisendi et al., 2006). Remarkably, heterozygous mutations of NPM are found in ∼35% of acute myeloid leukemias (AMLs), which makes NPM the most frequently mutated gene in AML (Falini et al., 2005). Despite their genetic heterogeneity (>20 mutations have been described), all AML mutations of NPM introduce a de novo nuclear export signal, which is responsible for the cytoplasmic accumulation of the mutated protein (Mariano et al., 2006).
The nature of the cancer alterations (overexpression and heterozygous mutations) initially led to NPM being proposed as a proto-oncogene. Mouse genetics experiments highlighted a different picture. Although inactivation of NPM in the germ line leads to embryonic lethality, studies in cultured NPM-null cells or mice carrying a single inactivated NPM allele provided evidence for a tumor-suppressor role. In particular, it was shown that: (1) NPM-null cells accumulate DNA damage (Colombo et al., 2005) and have increased centrosome numbers (Grisendi et al., 2005), which suggests genomic instability; and (2) heterozygous animals develop signs of myelodysplastic syndrome and display accelerated lymphomagenesis when crossed with μ-myc transgenic mice (Grisendi et al., 2005).
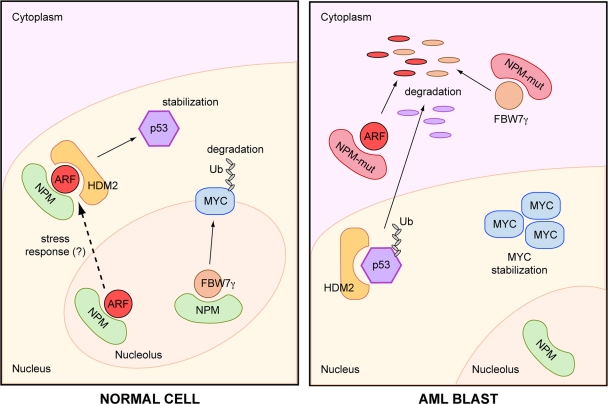
At the molecular level, the tumor suppressor activity of NPM seems to impinge directly and indirectly on the regulation of p53 (for review see Grisendi et al., 2006). One well-characterized mechanism involves the ARF tumor suppressor. In NPM-null cells, ARF loses its physiological localization in the nucleolus and becomes unstable, which suggests that NPM is critical for the proper localization and stability of ARF (Colombo et al., 2005). Notably, this function of NPM is lost for AML-associated NPM alleles (NPM-mut), which compete with wild-type NPM for ARF binding but target ARF to the cytoplasm, where it becomes more susceptible to degradation. This, in turn, leads to impaired ARF-dependent activation of p53 (Fig. 1; Colombo et al., 2006).
Figure 1.
Mutated NMP attenuates an oncosuppressor pathway and enhances an oncogenic one. Normal cell: NPM is mainly localized in the nucleolus and is required for nucleolar accumulation and stability of FBW7γ and ARF. This is relevant for the control of MYC turnover and provides an active pool of ARF ready to inactivate the HDM2-mediated p53 degradation in response to cellular stress. AML blast: NPM-mut is mainly localized to the cytoplasm and causes cytoplasmic delocalization and degradation of ARF and FBW7γ. As a consequence, HDM2 can induce ubiquitination/degradation of p53, and MYC accumulates and activates its target genes.
Does this settle the issue? Probably not, as one observation remains unexplained. AMLs carrying mutated NPM alleles display, by and large (∼85% of the cases), a normal karyotype, and show no apparent signs of genomic instability. How can this be reconciled with the fact that malignancies carrying inactivation of the ARF–p53 pathway are usually associated with genome instability and multiple cytogenetic abnormalities? In the general scheme of cancerogenesis, genomic instability is thought to represent the enabling condition that induces the emergence of additional genetic alterations needed to confer the selectable proliferative advantage to tumor cells. Of course, it cannot be excluded that as-yet-undetected point mutations (other than those affecting NPM) or minimal cytogenetic alterations are present in NPM-mutated AMLs. There is, however, one alternative possibility, i.e., that NPM-mut impinges both on tumor suppressor and oncogenic circuitries. Under this scenario, a mutant NPM allele might confer the quasi-entire repertoire of alterations needed for transformation, and there would be no selective pressure on leukemic clones displaying genomic instability (which might still be present but would simply be diluted within the total leukemic population).
In this issue, Bonetti et al. (see p. 19) describe one mechanism through which mutations of NPM might activate oncogenic pathways. They show that, in the absence of NPM or in the presence of NPM-mut, cells express increased levels of the MYC proto-oncogene. The critical player in this circuitry is FBW7γ, a nucleolar ubiquitin ligase previously implicated in the ubiquitination/degradation of MYC (Yada et al., 2004). The authors demonstrate that NPM interacts with FBW7γ and that, in the absence of NPM, FBW7γ loses its nucleolar localization and is rapidly degraded by the proteasome. As a consequence, ubiquitination of MYC is defective and the protein is stabilized. NPM-mut maintains the property of interacting with FBW7γ but delocalizes it to the cytoplasm, where it is degraded, thus leading to accumulation of MYC and increased MYC signaling (Fig. 1). Thus, mutations of NPM seem to simultaneously dampen a tumor-suppressor pathway (p53–ARF) and enhance an oncogenic one (MYC; Fig. 1). Alterations of NPM alone would therefore be sufficient to cause leukemia. To prove this point, one would need to show that the concomitant inactivation of ARF and activation of MYC are indeed necessary for the leukemogenic activity of NPM-mut, an issue that is still outstanding.
One remarkable feature of the system is that the mechanisms underlying the dual function of NPM-mut seem to adhere to the same molecular blueprint, i.e., that of “placing a critical cell regulator in the wrong place at the wrong time.” In the case of both ARF and FBW7γ (which, incidentally, is frequently mutated/deleted in human cancers and is a haploinsufficient tumor suppressor in mice; Minella and Clurman, 2005), the mislocalization of the proteins to the nucleoplasm or to the cytoplasm in NPM-null or NPM-mut–expressing cells, respectively, causes their inappropriate degradation. This provides fodder for new questions and further speculation. For instance, the degradation of ARF is, at least in part, ubiquitin independent and mediated by a noncanonical proteosomal pathway (the REGγ proteasome; Chen et al., 2007). Is a similar mechanism operational for FBW7γ as well? Moreover, the “physiological” function of NPM appears to be linked to ensuring the proper intracellular localization of ARF and FBW7γ (and possibly of other proteins). Is NPM a passive “reservoir” for these proteins? This seems unlikely, as NPM shuttles continuously between the nucleolus, the nucleoplasm, and the cytoplasm; this dynamic behavior is more readily compatible with some form of regulation of the trafficking of its interactors. In the case of the p53–ARF pathway, the hypothesis has been put forward that NPM might be a nucleolar sensor of oncogenic stress (Fig. 1). Under normal conditions, NPM sequesters ARF in the nucleolus. By sensing oncogene activation, or indeed, in other, more physiological conditions, NPM might traffic ARF to the nucleoplasm for activation of p53. Is this what is actually happening? And if so, how is it regulated? Are similar regulatory mechanisms involved in the NPM/FBW7γ circuitry? All these issues deserve further experimental attention.
Finally, it should be noted that FBW7γ is part of an SCF ubiquitination complex that targets other growth-promoting proteins, such as cyclin E and NOTCH, for degradation. Bonetti et al. (2008) did not investigate whether NPM and its mutants also regulate these proteins through the FBW7γ connection. This is an interesting issue, as both MYC and NOTCH regulate the self renewal of stem cells, and FBW7γ has been implicated in the control of stem cell quiescence (Thompson et al., 2008). These observations, together with those reported by Bonetti et al. (2008), lead us to question whether NPM is involved in the regulation of stem cell functions, an activity that could be critical for the selection of NPM mutations during leukemogenesis.
Acknowledgments
Work in the author's laboratory is supported by Associazione Italiana Ricerca sul Cancro (AIRC), European Community (VI Framework), Fondazione Monzino, Fondazione Ferrari, and Fondazione Cariplo.
Abbreviations used in this paper: AML, acute myeloid leukemia; NPM, nucleophosmin.
References
- Bonetti, P., T. Davoli, C. Sironi, B. Amati, P.G. Pelicci, and E. Colombo. 2008. Nucleophosmin and its AML-associated mutant regulate c-Myc turnover through Fbw7γ. J. Cell Biol. 182:19–26. [DOI] [PMC free article] [PubMed]
- Chen, X., L.F. Barton, Y. Chi, B.E. Clurman, and J.M. Roberts. 2007. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Mol. Cell. 26:843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo, E., P. Bonetti, E. Lazzerini Denchi, P. Martinelli, R. Zamponi, J.C. Marine, K. Helin, B. Falini, and P.G. Pelicci. 2005. Nucleophosmin is required for DNA integrity and p19Arf protein stability. Mol. Cell. Biol. 25:8874–8886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo, E., P. Martinelli, R. Zamponi, D.C. Shing, P. Bonetti, L. Luzi, S. Volorio, L. Bernard, G. Pruneri, M. Alcalay, and P.G. Pelicci. 2006. Delocalization and destabilization of the Arf tumor suppressor by the leukemia-associated NPM mutant. Cancer Res. 66:3044–3050. [DOI] [PubMed] [Google Scholar]
- Falini, B., C. Mecucci, E. Tiacci, M. Alcalay, R. Rosati, L. Pasqualucci, R. La Starza, D. Diverio, E. Colombo, A. Santucci, et al. 2005. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl. J. Med. 352:254–266. [DOI] [PubMed] [Google Scholar]
- Grisendi, S., R. Bernardi, M. Rossi, K. Cheng, L. Khandker, K. Manova, and P.P. Pandolfi. 2005. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 437:147–153. [DOI] [PubMed] [Google Scholar]
- Grisendi, S., C. Mecucci, B. Falini, and P.P. Pandolfi. 2006. Nucleophosmin and cancer. Nat. Rev. Cancer. 6:493–505. [DOI] [PubMed] [Google Scholar]
- Mariano, A.R., E. Colombo, L. Luzi, P. Martinelli, S. Volorio, L. Bernard, N. Meani, R. Bergomas, M. Alcalay, and P.G. Pelicci. 2006. Cytoplasmic localization of NPM in myeloid leukemias is dictated by gain-of-function mutations that create a functional nuclear export signal. Oncogene. 25:4376–4380. [DOI] [PubMed] [Google Scholar]
- Minella, A.C., and B.E. Clurman. 2005. Mechanisms of tumor suppression by the SCF(Fbw7). Cell Cycle. 4:1356–1359. [DOI] [PubMed] [Google Scholar]
- Thompson, B.J., V. Jankovic, J. Gao, S. Buonamici, A. Vest, J.M. Lee, J. Zavadil, S.D. Nimer, and I. Aifantis. 2008. Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J. Exp. Med. 205:1395–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yada, M., S. Hatakeyama, T. Kamura, M. Nishiyama, R. Tsunematsu, H. Imaki, N. Ishida, F. Okumura, K. Nakayama, and K.I. Nakayama. 2004. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 23:2116–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]