Abstract
Actin dynamics provide the driving force for many cellular processes including motility and endocytosis. Among the central cytoskeletal regulators are actin-depolymerizing factor (ADF)/cofilin, which depolymerizes actin filaments, and twinfilin, which sequesters actin monomers and caps filament barbed ends. Both interact with actin through an ADF homology (ADF-H) domain, which is also found in several other actin-binding proteins. However, in the absence of an atomic structure for the ADF-H domain in complex with actin, the mechanism by which these proteins interact with actin has remained unknown. Here, we present the crystal structure of twinfilin's C-terminal ADF-H domain in complex with an actin monomer. This domain binds between actin subdomains 1 and 3 through an interface that is conserved among ADF-H domain proteins. Based on this structure, we suggest a mechanism by which ADF/cofilin and twinfilin inhibit nucleotide exchange of actin monomers and present a model for how ADF/cofilin induces filament depolymerization by weakening intrafilament interactions.
Introduction
Polymerization of actin filaments against membranes produces pushing forces that are required for various cellular processes such as motility, morphogenesis, and endocytosis (Pollard and Borisy, 2003; Kaksonen et al., 2006). Despite the large number of proteins regulating actin dynamics, many of them interact with actin through a relatively small number of protein domains. Among the central actin-binding domains is the actin-depolymerizing factor homology (ADF-H) domain, which occurs in five functionally distinct classes of proteins: ADF/cofilin, twinfilin, Abp1/drebrin, coactosin, and glia maturation factor (Paavilainen et al., 2007).
The founding member of this family, ADF/cofilin, binds both monomeric and filamentous actin, preferably in the ADP-bound form, and induces a structural rearrangement in the actin filament that leads to its disassembly. When bound to an actin monomer, ADF/cofilin inhibits spontaneous nucleotide exchange (Carlier et al., 1997; Bamburg, 1999; Andrianantoandro and Pollard, 2006). In cells, ADF/cofilin plays an essential role in various processes by promoting disassembly of aged actin filaments (Okreglak and Drubin, 2007). In contrast to ADF/cofilin, which consists of a single ADF-H domain, twinfilin is composed of two ADF-H domains separated by a short linker region (Paavilainen et al., 2004). Twinfilin binds ADP-actin monomers and filament barbed ends with high affinity, and prevents monomer assembly into filament ends (Ojala et al., 2002; Helfer et al., 2006). In addition, yeast twinfilin induces filament severing at a low pH (Moseley et al., 2006). Biochemical studies suggested that during barbed-end capping, twinfilin's N-terminal ADF-H domain interacts with the terminal actin subunit, whereas the C-terminal ADF-H domain binds to the side of an actin filament through a similar mechanism to that of ADF/cofilin (Paavilainen et al., 2007). The exact functions of the Abp1/drebrin, coactosin, and glia maturation factor are less well understood, although also these proteins are linked to regulation of actin dynamics (de Hostos et al., 1993; Quintero-Monzon et al., 2005; Ikeda et al., 2006).
Although the biochemical activities and cellular functions of ADF-H domain proteins are rapidly being uncovered, the structure of an ADF-H domain in complex with actin has not been reported. Indirect structural methods have provided controversial results, and even the binding site of this domain on actin is not known (Wriggers et al., 1998; Kamal et al., 2007). Consequently, the structural mechanisms by which twinfilin and ADF/cofilin inhibit nucleotide exchange on actin monomers and how ADF/cofilin induces filament depolymerization/severing are unknown.
Results and discussion
To reveal how ADF-H domain proteins interact with actin, we set out to crystallize the C-terminal ADF-H domain of twinfilin corresponding to residues 176–316 (hereafter termed Twf-C) with ATP– and ADP–G-actin. Similarly to ADF/cofilins, isolated Twf-C binds actin monomers and filaments, preferring ADP-actin, and induces filament depolymerization, although with a lower efficiency. The structure of Twf-C is very similar to that of ADF/cofilin (Cα rmsd 2.0 Å for 130 superposed residues of yeast cofilin), and it interacts with actin through a very similar interface (Paavilainen et al., 2007). Thus, Twf-C also serves as a good model for studying how ADF/cofilin interacts with actin.
Structure of Twf-C in complex with ATP–G-actin
Despite numerous attempts, we did not obtain crystals of Twf-C in complex with ADP-G-actin. However, crystals of the Twf-C/ATP–G-actin complex were obtained from 15% PEG3350, pH 9.0, and the structure was determined by molecular replacement. The crystals contained one copy of Twf-C and ATP–G-actin, and the final model was refined to a resolution of 2.55 Å (Fig. 1 and Table I). Comparison of Twf-C structure in solution (Paavilainen et al., 2007) and in complex with G-actin demonstrates that the ADF-H domain does not undergo major conformational changes upon binding to actin monomer (Cα rmsd 1.5 Å for 139 superposed residues). The only significant structural change was observed in the two N-terminal residues of Twf-C (residues 176–177), which are part of a flexible extension in most ADF-H domain structures without actin (Paavilainen et al., 2002, 2007; Hellman et al., 2004; Quintero-Monzon et al., 2005; Andrianantoandro and Pollard, 2006), but become ordered in complex with actin and form an important part of the interaction surface (see following paragraph).
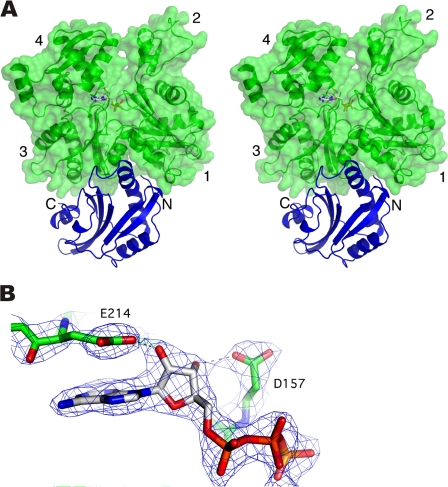
Figure 1.
Crystal structure of the Twf-C–actin monomer complex at 2.55 Å resolution. (A) Twf-C (blue) binds between the actin (green) subdomains 1 and 3, and buries a surface area of ∼1,200 Å2. The ATP molecule bound to actin is shown in atom colors. (B) The σA weighted 2 mFo-DFc electron density map around the ATP molecule contoured at 1 σ.
Table I. X-ray data collection and refinement statistics.
| Data collection | TwfC-actin |
|---|---|
| Space group | P212121 |
| Unit cell parameters | a = 52.8; b = 73.0; c = 168.9 |
| Resolution range (Å) | 2.55–42.8 |
| Highest resolution shell (Å) | 2.55–2.61 |
| Measured reflections | 83,041 (4,553) |
| Unique reflections | 26,755 (1,318) |
| Redundancy | 3.1 (3.5) |
| Completeness (%) | 99.3 (99.8) |
| Mean I/σ | 9.6 (2.3) |
| Rsym (%) | 13.1 |
| Rcryst | 20.8 (30.4) |
| Rfree | 27.9 (37.7) |
| No. of protein atoms | 4,056 |
| No. of water molecules | 123 |
| Wilson B-value (Å2) | 39.3 |
| Mean B factors (Å2) | |
| Twf-C | 43.6 |
| Actin | 49.1 |
| Solvent | 41.9 |
| Ramachandran (%) | |
| Most favored | 89.6 |
| Additionally allowed | 10.0 |
| Generously allowed | 0.4 |
| Disallowed | 0.0 |
| rms deviations | |
| Bond lengths (Å) | 0.012 |
| Bond angles (o) | 1.5 |
Values in parentheses are for the highest resolution bin.
The mechanism of ADF-H domain–actin interaction
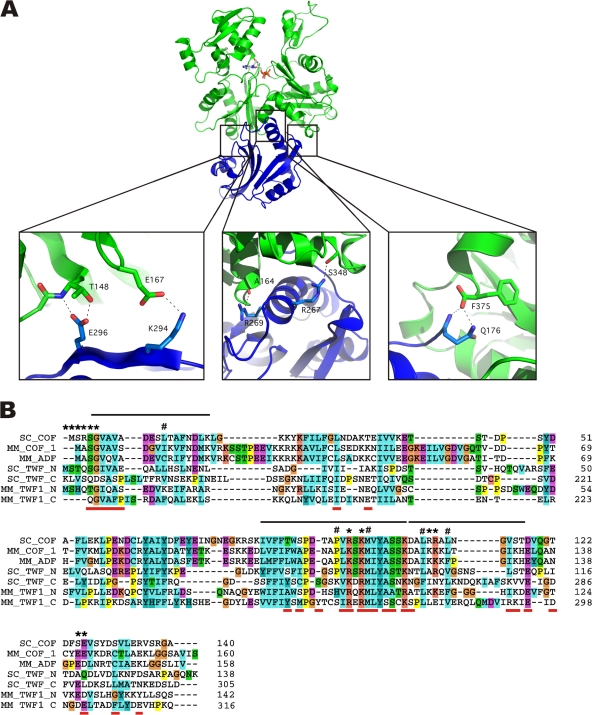
Twf-C binds to a groove between actin subdomains 1 and 3 through an interface that buries a surface area of ∼1200 Å2 (Fig. 1 A). Three major sites of interaction can be distinguished: (1) the N-terminal extension of the domain (twinfilin residues 176–181); (2) the long α-helix (twinfilin residues 266–274); and (3) the region before the C-terminal helix of this domain (twinfilin residues 294–302; Fig. 2 A). Within these regions, the most obvious contacts are made between residues Q176 of twinfilin (Twf) and the C-terminal F375 of actin, R267 (Twf) and S348 (actin), R269 (Twf) and A144 (actin), S273 (Twf) and Y143 (actin), K276 (Twf) and T148 (actin), K294 (Twf) and E167 (actin), and E296 (Twf) and T148 (actin; Fig. 2). In addition, several residues are involved in hydrophobic contacts across the interface. These include V178 (Twf) and L346, L349, T351, F352, M355 (actin), F180 (Twf) and L349, T351 (actin), I266 (Twf) and E334, I341, I345 (actin), M270 (Twf) and A144, G342, I346, L346 (actin), and L271 (Twf) and L349 (actin). Additionally, 10 water molecules are found at the interface. With the exception of Q176, which is the first residue in the C-terminal ADF-H domain of twinfilin and can make a hydrogen bond with actin through its main-chain amide group and S274, these residues are highly conserved in ADF/cofilins and in both twinfilin domains (Fig. 2 B). Previous mutagenesis and biochemical studies revealed that these regions are critical for actin interactions in both ADF/cofilin and twinfilin (Lappalainen et al., 1997; Guan et al., 2002; Paavilainen et al., 2002, 2007; Grintsevich et al., 2008). Furthermore, recent cross-linking studies suggested that these regions are important for ADF/cofilin interactions in actin (Grintsevich et al., 2008). Thus, the Twf-C/G-actin structure provides a good structural model for the G-actin–bound state of ADF/cofilin (Fig. 3).
Figure 2.
Conservation of the actin monomer binding site in ADF/cofilins and twinfilins. (A) Three major sites of interaction are present in the Twf-C–G-actin structure: (left) The N-terminal extension of Twf-C, (middle) the long α-helix, and (right) the loop before the C-terminal helix. Close-up figures illustrate some of the major contacts observed in the structure. (B) A structural sequence alignment between Twf-C, Twf-N, and ADF/cofilin. The residues shown to be important for G-actin interactions by mutagenesis are indicated by asterisks. Residues identified in a synchrotron footprinting study as G-actin–interacting residues in ADF/cofilin are indicated by hash marks. Interacting peptides from the same study are shown as black lines. Interface residues identified from our crystal structure are displayed as red lines below the sequences.
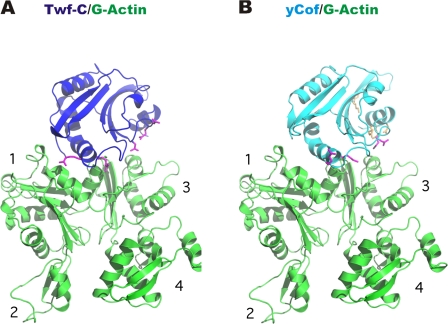
Figure 3.
Twf-C and ADF/cofilin bind G-actin through a conserved mechanism. (A) Structure of the Twf-C–G-actin complex and (B) a model of yeast cofilin (Fedorov et al., 1997) bound to G-actin in the same orientation show that both proteins use a similar binding surface for G-actin. Residues shown to be important for G-actin binding by mutagenesis in Twf-C and yeast cofilin are displayed as magenta sticks, and residues important for F-actin interactions in yeast cofilin are shown in orange.
The same regions that are important for G-actin binding in ADF/cofilin and twinfilin domains are also critical for actin interactions in coactosin and Abp1/drebrin (Quintero-Monzon et al., 2005; Dai et al., 2006). However, these regions, and especially the long α-helix, are less conserved in coactosin and Abp1, which bind F-actin with relatively low affinity and do not interact with G-actin or induce filament disassembly (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200803100/DC1). Thus, we propose that the inability of the long α-helix of these proteins to interact tightly with the groove between actin subdomains 1 and 3 may be responsible for the lack of G-actin binding and the F-actin disassembly activities of coactosin and Abp1.
In 7-chloro-4-nitrobenz-2-oxa-1,3-diazole (NBD)-actin and pyrenyl-actin, the fluorophores are attached to actin residues K373 and C374, which are located at the ADF-H domain binding interface. This may provide an explanation for why ADF/cofilin binding induces a change in the fluorescence of NBD–G-actin (Carlier et al., 1997), and twinfilin binding induces a change in fluorescence of both NBD–G-actin and pyrenyl–G-actin (Ojala et al., 2002; Falck et al., 2004). Furthermore, phosphorylation of an N-terminal serine results in inhibition of both the G- and F-actin–binding activity of ADF/cofilins (Bamburg, 1999). In our crystal structure, the corresponding residue of Twf-C (Q176) is located at the binding interface, which provides a good explanation for why phosphorylation of this residue in ADF/cofilin inhibits its interaction with actin.
Comparison of the G-actin interactions of ADF-H and gelsolin domains
Comparison of the Twf-C–G-actin complex to the structures of other central actin-binding domains in complex with actin provides further evidence for the model in which the majority of actin-regulating proteins bind to a “hot spot” groove on the actin monomer (Dominguez, 2004). Similarly to gelsolin and WASP homology 2 (WH2) domains, the major protein–protein contact in the ADF-H domain involves a long α-helix, which interacts with the hydrophobic groove located between actin subdomains 1 and 3 (Fig. 4 A; McLaughlin et al., 1993; Burtnick et al., 2004; Hertzog et al., 2004; Chereau et al., 2005).
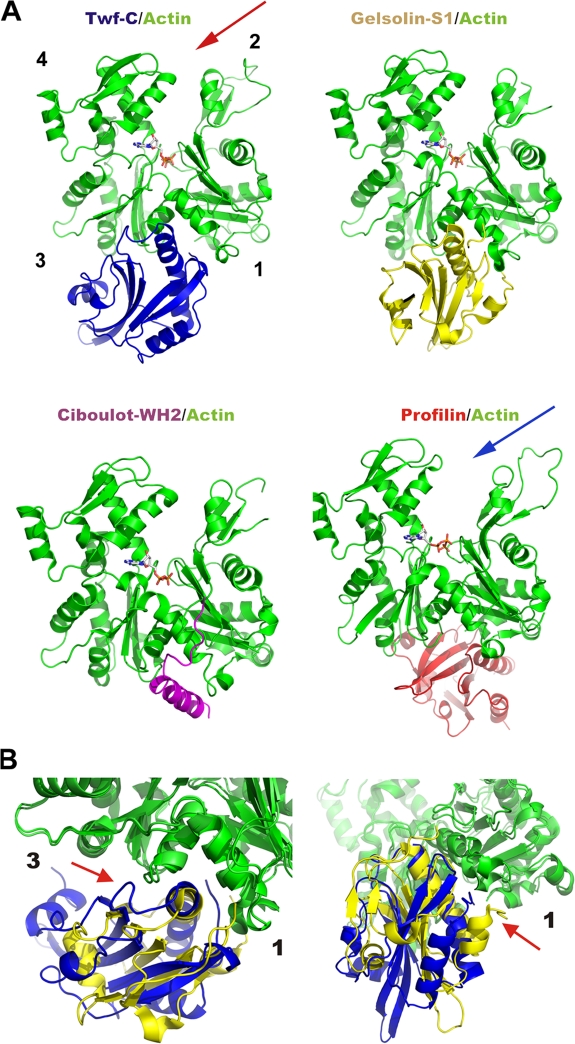
Figure 4.
Comparison of the ADF-H domain–actin complex with other conserved actin-binding domains in complex with G-actin. (A) Twf-C binds to the “hot spot” between actin subdomains 1 and 3 similarly to gelsolin segment 1 (S1) and the WH2 domain of ciboulot. These three proteins inhibit the nucleotide exchange on the actin monomer and keep the cleft between actin subdomains 2 and 4 in a “closed” conformation (red arrow). Bovine profilin binds “behind” the hydrophobic cleft between actin subdomain 1 and 3. However, profilin appears to maintain the actin monomer in an “open” state (blue arrow) and promotes nucleotide exchange. (B) Comparison of G-actin interactions of gelsolin S1 and Twf-C. Gelsolin S1 (McLaughlin et al., 1993) is shown in yellow and Twf-C in blue. The most significant differences in the actin interactions are indicated by red arrows. These are: interaction of the loop before the C-terminal α-helix of Twf-C with actin subdomain 3 (left) and different interaction sites of N-terminal extensions of Twf-C and gelsolin S1 in actin (right).
However, although ADF-H domain and gelsolin segment-1 are structurally related, there are significant differences in the mechanisms by which they interact with actin. In both domains, the N-terminal region before the first α-helix is involved in actin binding, but these regions interact with different faces of helix-4 of actin. Although the loop before the C-terminal α-helix plays a central role in G-actin binding in the ADF-H domain, the corresponding region in gelsolin segment-1 does not contact actin (Fig. 4 B). Finally, although the long α-helix forms the major actin-binding site in both domains and incorporates to the groove between actin subdomains 1 and 3 in a nearly identical orientation, the actual contacts between this helix and actin are relatively poorly conserved between ADF-H and gelsolin domains. For example, the two basic residues (R267 and R269) that make important contacts with actin in the Twf-C/ATP–G-actin structure (Fig. 2), which have been shown to be critical for actin interactions in ADF/cofilins and both twinfilin domains (Lappalainen et al., 1997; Paavilainen et al., 2002, 2007), are not conserved in gelsolin domains.
Inhibition of nucleotide exchange by ADF-H domain proteins
Twinfilin, ADF/cofilin, gelsolin, and WH2 domain proteins inhibit spontaneous nucleotide exchange when bound to an actin monomer (Tellam, 1986; Bamburg, 1999; Hertzog et al., 2004; Paavilainen et al., 2004). Also, the isolated C-terminal ADF-H domain of twinfilin binds G-actin with high affinity and efficiently inhibits G-actin nucleotide exchange (Fig. S1). In contrast, most profilins promote nucleotide exchange in actin monomers (Witke, 2004). Comparison of the actin conformation in complex with Twf-C, profilin, gelsolin-S1, and ciboulot WH2 domains (all of which were crystallized without DNase I bound to subdomains 2 and 4) reveals that in complexes that inhibit nucleotide exchange, the cleft between actin subdomains 2 and 4 is in a “closed” state. In contrast, in the profilin–actin complex, the cleft between actin subdomains 2 and 4 is “open” (Schutt et al., 1993), which may allow more rapid exchange of the nucleotide (Fig. 4 A). Thus, the differences in how these proteins interact with the groove between actin subdomains 1 and 3 may induce conformational changes in the actin molecule that control the accessibility of the nucleotide to the solvent through the cleft between subdomains 2 and 4.
Interaction of Twf-C and ADF/cofilins with actin filaments
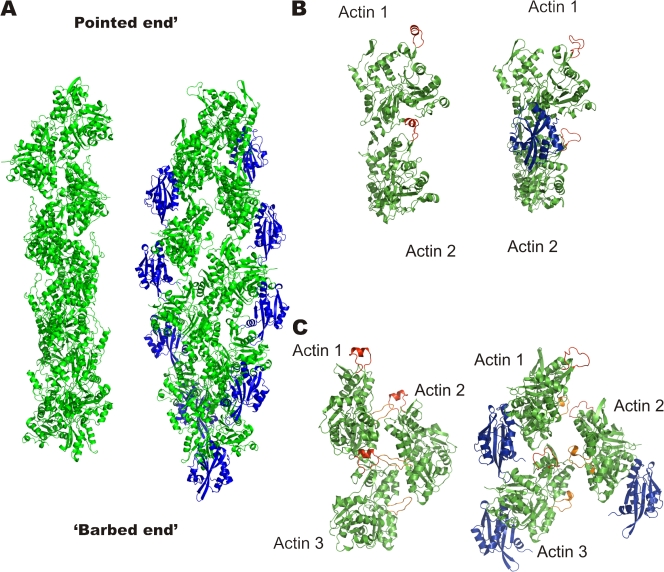
ADF/cofilin binds to actin filaments in a cooperative fashion and induces filament disassembly, most likely via weakening of intramolecular contacts in the actin filament (Bamburg, 1999). Because Twf-C also binds actin filaments (although with lower affinity than ADF/cofilin) and induces filament disassembly (Paavilainen et al., 2007), we decided to build a model for the Twf-C–bound actin filaments. We first attempted to overlay the actin monomer from our crystal structure with an actin monomer from two different actin filament models. Neither the original fiber diffraction–based model of naked actin filaments (Holmes et al., 1990) nor the EM-based model of an ADF/cofilin-decorated actin filament (Galkin et al., 2001) produced a good fit with the Twf-C–G-actin complex. Although the conformation of the actin monomer in both filament models is nearly identical to that in our crystal structure, the orientation in the original Holmes et al. (1990) model is such that the bound ADF-H domain clashes with the next actin monomer in the strand. A similar problem occurs with the Galkin et al. (2001), model; the ADF-H domain in their model is in a slightly different orientation compared with our crystal structure, which leads to severe clashes with the next actin monomer.
Cryo-EM analyses revealed that ADF/cofilin binding affects the actin filament conformation by stabilizing a filament state with a mean twist of 162° (McGough et al., 1997; Galkin et al., 2001). We fitted the high-resolution Twf-C–G-actin structure into the experimental EM-based electron density map from Galkin et al., (2003). Automatic docking procedure resulted in a good fit with the experimental map, resulting in a new ADF-H domain–decorated actin filament model with a mean rotational angle of 162.2° and a mean translation of 27.7 Å (Fig. 5). Additionally, a good fit with the Twf-C–G-actin structure was obtained with the latest fiber diffraction–based filament model (Holmes et al., 2003), which has significant domain movements compared with the G-actin structure. In the model, the so-called D-loop of actin (residues 38–52) forms a helix, which binds between subdomains 1 and 3 of the next actin monomer. Fitting our structure into the EM electron density map (Galkin et al., 2003) and the filament model (Holmes et al., 2003) resulted in two similar ADF-H–decorated filament models with 162° and 167° twists. The 162° model (Fig. 5 A) may represent the optimal filament for ADF/cofilin binding, whereas the 167° model (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200803100/DC1) may correspond to an initial binding mode for ADF/cofilin.
Figure 5.
A hypothetical model for an ADF-H domain decorated actin filament. (A) A model of an ADF-H domain–decorated actin filament obtained by docking the Twf-C–G-actin structure in the 23-Å electron density map of an ADF/cofilin-decorated actin filament (Galkin et al., 2003). (B) Binding of Twf-C (and ADF/cofilin) in this model results in a large structural change of actin subdomain 2, where the so-called D-loop (shown in red) moves ∼17 Å away from the actin hot spot cleft. (C) In the model, the structural rearrangement of actin subdomain 2 also affects the interaction between the two actin strands by weakening contacts involving the so-called actin “hydrophobic loop” (colored in orange). Together, these changes in inter- and cross-filament interactions could contribute to weakening of the actin filament and lead to filament depolymerization by Twf-C and ADF/cofilin.
Comparison of the filament models in the presence and absence of Twf-C suggests that interaction of the long α-helix of Twf-C with the groove between actin subdomains 1 and 3 forces the D-loop of the adjacent monomer to move ∼17 Å away from the actin hot spot cleft (Fig. 5 B). Replacing Twf-C with yeast cofilin in the model suggests that the actin filament–binding site of cofilin buries an area of ∼1500 Å2. ADF/cofilin residues R80, K82, E134, R135, and R138, which were previously shown to be important for F-actin binding by mutagenesis (Lappalainen et al., 1997), are located at the interface (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200803100/DC1). This provides further evidence that the ADF-H domain–binding mode observed in our crystal structure is also similar to the filament-bound form of ADF/cofilin. However, this region is not conserved between ADF/cofilin and Twf-C, which provides a possible explanation for the weaker F-actin–binding and disassembly activities of Twf-C as compared with ADF/cofilins.
The model presented in Fig. 5 is consistent with recent proteolysis, Förster resonance energy transfer (FRET), and cross-linking experiments demonstrating that ADF/cofilin binding to actin filament results in a structural rearrangement of actin subdomain 2 and exposure of the D-loop (Bobkov et al., 2002; Muhlrad et al., 2004). It should be noted that even though the actin subdomain 2 is involved in a crystal contact in our crystals, the D-loop does not contribute to these contacts. We propose that rearrangement of the D-loop upon ADF-H domain binding may result in weakening of the interfilament contacts between successive actin monomers. Additionally, the “hydrophobic loop” of actin (residues 262–274), which mediates cross-filament interactions in the Holmes model, has been shown to be important for filament growth and stability (Shvetsov et al., 2008). In our model, the structural rearrangement of the actin subdomain 2 in the filament decorated with the ADF-H domain (Fig. 5 C and Videos 1 and 2, available at http://www.jcb.org/cgi/content/full/jcb.200803100/DC1) appears to also weaken these cross-filament contacts, which may result in filament disassembly (Carlier et al., 1997; Andrianantoandro and Pollard, 2006).
In conclusion, we show that ADF-H domains bind between actin subdomains 1 and 3 using a similar insertion of an α-helix into the hydrophobic cleft of actin as described previously for gelsolin and WH2 domains. Binding of ADF-H domain appears to lock the cleft between actin subdomains 2 and 4 in a closed conformation, which may provide a structural explanation for how ADF/cofilin and twinfilin inhibit nucleotide exchange in actin monomers. We also propose a model for how Twf-C and ADF/cofilin induce filament disassembly through weakening of both longitudinal and lateral contacts within the actin filament. It is important to note that the crystal structure used in our modeling was from the ADF-H domain/ATP–G-actin complex, whereas at least ADF/cofilin binds ADP-actin with much higher affinity than ATP-actin (Carlier et al., 1997). Although the structures of ATP– and ADP–G-actin were found to be similar to each other (Otterbein et al., 2001; Rould et al., 2006), further studies will be required to reveal the structural changes that occur in F-actin upon nucleotide hydrolysis and how they affect the interactions with ADF-H domain proteins.
Materials and methods
Cloning, protein expression, purification, and crystallization
The mouse twinfilin-1176-316 (Twf-C) construct was cloned into NcoI–HindIII sites of a pHAT2 vector. The Twf-C protein was expressed in Escherichia coli BL21(DE3) cells and purified by using a chelating Sepharose column loaded with Ni2+ ions, followed by gel filtration with a Superdex 75 10/60 column (GE Healthcare; Paavilainen et al., 2007). Rabbit muscle actin was purified and labeled with NBD as described previously (Ojala et al., 2002). Protein concentrations were determined by using the calculated extinction coefficients at 280 nm with a diode array spectrophotometer (8452A; Hewlett-Packard). Twf-C was mixed with ATP–G-actin, and the complex was purified by gel filtration with a Superdex 75 10/60 column. The complex was then concentrated to 8 mg/ml in modified G buffer (10 mM Tris, pH 7.5, 50 mM NaCl, 0.2 mM ATP, 0.2 mM DTT, and 0.2 mM CaCl2). Crystals of the complex were grown in hanging drops by mixing 1 μl of Twf-C–G-actin complex with 1 μl of precipitant solution composed of 0.1M Hepes–CHES–citric acid, pH 9.0, 15% PEG3350, and 0.1 M guanidium hydrochloride. Crystals were then transferred to a precipitant solution, supplemented with 30% glycerol, and frozen in a stream of liquid nitrogen at 100°K.
Data collection and structure solution
The crystals belonged to the space group P212121 with unit cell parameters a = 52.8 Å; b = 73.0 Å; and c = 168.9 Å. A dataset was collected on the ID23-1 beamline of the European Synchrotron Radiation Facility in Grenoble, France. Diffraction data were integrated and scaled with the program XDS (Table I). The structure of the Twf-C–G-actin complex was solved by molecular replacement using data to 2.55 Å and structures 2hd7 (Paavilainen et al., 2007) and 2a42 (Chereau et al., 2005) as search models in the program PHASER (McCoy, 2007), followed by several rounds of manual rebuilding and restrained refinement with programs COOT and REFMAC5 (Table I). Water molecules were added by ARP/WARP. The structure was validated with the MolProbity server. The coordinates of the model were deposited in the Protein Data Bank (PDB accession code 3DAW).
Preparation of the ADF-H–decorated F-actin models
To obtain a model of the Twf-C/F-actin with a 167° twist, the crystal structure of the actin–Twf-C complex was superimposed individually on five monomers of the Holmes F-actin model (coordinates from http://www.mpimf-heidelberg.mpg.de/∼holmes/; Holmes et al., 2003) to generate a model of the decorated filament. The Twf-C molecules were first placed onto the Holmes filament model and the actin monomers were then morphed to the G-actin–like conformation of the crystal structure, using the torsion angle morph as implemented in LSQMAN (Kleywegt, 1996). To prepare the Twf-C–F-actin model with a 162° twist, the structure was docked to a 23-Å electron density map (provided by E. Egelman, University of Virginia, Charlottesville, VA; Galkin et al., 2003) using correlation-based docking in the program Situs 2.3 (Chacon and Wriggers, 2002). Nine molecules forming a continuous filament were located without imposing any helical symmetry constraints in the docking. The molecules were superposed on the next molecule in the filament to determine the rotation matrix and translation between the two monomers. The mean rotation angle was 162.2° and the mean translation was 27.7 Å.
Biochemical experiments
NBD–G-actin binding and nucleotide exchange assays were performed as described previously (Ojala et al., 2002).
Online supplemental material
Fig. S1 shows that Twf-C interacts with actin monomers with high affinity and inhibits the nucleotide exchange of actin monomers. Fig. S2 presents a hypothetical model for an ADF-H domain–decorated actin filament with a 167° twist. Fig. S3 shows a model for the ADF–cofilin interaction with F-actin. Videos 1 and 2 propose a model for how ADF-H domain binding to actin filaments causes a rearrangement in actin subdomain 2. Table S1 describes the actin-binding activities of the different ADF-H domain–containing proteins. Video 1 shows a side view of ADF-H domain binding to the actin filament. Video 2 shows a top view of ADF-H domain binding to the actin filament. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200803100/DC1.
Acknowledgments
We acknowledge the European Synchrotron Radiation Facility for provision of synchrotron radiation facilities and Dr. Joanne McCarthy for assistance in using the beamline ID23-1. Edward Egelman is acknowledged for kindly providing the cofilin/F-actin electron density map. Jussi Jäntti, Tom Pollard, and Maria Vartiainen are acknowledged for critical comments on the manuscript.
E. Oksanen acknowledges funding from the Informational and Structural Biology graduate school. Work in P. Lappalainen's and A. Goldman's laboratories was funded by Biocentrum Helsinki, the Sigrid Jusélius foundation, and the Academy of Finland.
V.O. Paavilainen's present address is Department of Cellular and Molecular Pharmacology, University of California, San Francisco, San Francisco, CA 94158.
Abbreviations used in this paper: ADF-H, actin-depolymerizing factor homology; NBD, 7-chloro-4-nitrobenz-2-oxa-1,3-diazole; WH2, WASP homology 2.
References
- Andrianantoandro, E., and T.D. Pollard. 2006. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell. 24:13–23. [DOI] [PubMed] [Google Scholar]
- Bamburg, J.R. 1999. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 15:185–230. [DOI] [PubMed] [Google Scholar]
- Bobkov, A.A., A. Muhlrad, K. Kokabi, S. Vorobiev, S.C. Almo, and E. Reisler. 2002. Structural effects of cofilin on longitudinal contacts in F-actin. J. Mol. Biol. 323:739–750. [DOI] [PubMed] [Google Scholar]
- Burtnick, L.D., D. Urosev, E. Irobi, K. Narayan, and R.C. Robinson. 2004. Structure of the N-terminal half of gelsolin bound to actin: roles in severing, apoptosis and FAF. EMBO J. 23:2713–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier, M.F., V. Laurent, J. Santolini, R. Melki, D. Didry, G.X. Xia, Y. Hong, N.H. Chua, and D. Pantaloni. 1997. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 136:1307–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacon, P., and W. Wriggers. 2002. Multi-resolution contour-based fitting of macromolecular structures. J. Mol. Biol. 317:375–384. [DOI] [PubMed] [Google Scholar]
- Chereau, D., F. Kerff, P. Graceffa, Z. Grabarek, K. Langsetmo, and R. Dominguez. 2005. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc. Natl. Acad. Sci. USA. 102:16644–16649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, H., W. Huang, J. Xu, B. Yao, S. Xiong, H. Ding, Y. Tang, H. Liu, J. Wu, and Y. Shi. 2006. Binding model of human coactosin-like protein with filament actin revealed by mutagenesis. Biochim. Biophys. Acta. 1764:1688–1700. [DOI] [PubMed] [Google Scholar]
- de Hostos, E.L., B. Bradtke, F. Lottspeich, and G. Gerisch. 1993. Coactosin, a 17 kDa F-actin binding protein from Dictyostelium discoideum. Cell Motil. Cytoskeleton. 26:181–191. [DOI] [PubMed] [Google Scholar]
- Dominguez, R. 2004. Actin-binding proteins–a unifying hypothesis. Trends Biochem. Sci. 29:572–578. [DOI] [PubMed] [Google Scholar]
- Falck, S., V.O. Paavilainen, M.A. Wear, J.G. Grossmann, J.A. Cooper, and P. Lappalainen. 2004. Biological role and structural mechanism of twinfilin – capping protein interaction. EMBO J. 23:3010–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov, A.A., P. Lappalainen, E.V. Fedorov, D.G. Drubin, and S.C. Almo. 1997. Structure determination of yeast cofilin. Nat. Struct. Biol. 4:366–369. [DOI] [PubMed] [Google Scholar]
- Galkin, V.E., A. Orlova, N. Lukoyanova, W. Wriggers, and E.H. Egelman. 2001. Actin depolymerizing factor stabilizes an existing state of F-actin and can change the tilt of F-actin subunits. J. Cell Biol. 153:75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin, V.E., A. Orlova, M.S. VanLoock, A. Shvetsov, E. Reisler, and E.H. Egelman. 2003. ADF/cofilin use an intrinsic mode of F-actin instability to disrupt actin filaments. J. Cell Biol. 163:1057–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grintsevich, E.E., S.A. Benchaar, D. Warshaviak, P. Boontheung, F. Halgand, J.P. Whitelegge, K.F. Faull, R.R. Loo, D. Sept, J.A. Loo, and E. Reisler. 2008. Mapping the cofilin binding site on yeast G-actin by chemical cross-linking. J. Mol. Biol. 377:395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, J.Q., S. Vorobiev, S.C. Almo, and M.R. Chance. 2002. Mapping the G-actin binding surface of cofilin using synchrotron protein footprinting. Biochemistry. 41:5765–5775. [DOI] [PubMed] [Google Scholar]
- Helfer, E., E.M. Nevalainen, P. Naumanen, S. Romero, D. Didry, D. Pantaloni, P. Lappalainen, and M.F. Carlier. 2006. Mammalian twinfilin sequesters ADP-G-actin and caps filament barbed ends: implications in motility. EMBO J. 25:1184–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman, M., V.O. Paavilainen, P. Naumanen, P. Lappalainen, A. Annila, and P. Permi. 2004. Solution structure of coactosin reveals structural homology to ADF/cofilin family proteins. FEBS Lett. 576:91–96. [DOI] [PubMed] [Google Scholar]
- Hertzog, M., C. van Heijenoort, D. Didry, M. Gaudier, J. Coutant, B. Gigant, G. Didelot, T. Préat, M. Knossow, E. Guittet, and M.F. Carlier. 2004. The beta-thymosin/WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell. 117:611–623. [DOI] [PubMed] [Google Scholar]
- Holmes, K.C., D. Popp, W. Gebhard, and W. Kabsch. 1990. Atomic model of the actin filament. Nature. 347:44–49. [DOI] [PubMed] [Google Scholar]
- Holmes, K.C., I. Angert, F.J. Kull, W. Jahn, and R.R. Schroder. 2003. Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature. 425:423–427. [DOI] [PubMed] [Google Scholar]
- Ikeda, K., R.K. Kundu, S. Ikeda, M. Kobara, H. Matsubara, and T. Quertermous. 2006. Glia maturation factor-gamma is preferentially expressed in microvascular endothelial and inflammatory cells and modulates actin cytoskeleton reorganization. Circ. Res. 99:424–433. [DOI] [PubMed] [Google Scholar]
- Kaksonen, M., C.P. Toret, and D.G. Drubin. 2006. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 7:404–414. [DOI] [PubMed] [Google Scholar]
- Kamal, J.K., S.A. Benchaar, K. Takamoto, E. Reisler, and M.R. Chance. 2007. Three-dimensional structure of cofilin bound to monomeric actin derived by structural mass spectrometry data. Proc. Natl. Acad. Sci. USA. 104:7910–7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt, G.J. 1996. Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr. D Biol. Crystallogr. 52:842–857. [DOI] [PubMed] [Google Scholar]
- Lappalainen, P., E.V. Fedorov, A.A. Fedorov, S.C. Almo, and D.G. Drubin. 1997. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J. 16:5520–5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy, A.J. 2007. Phaser crystallographic software. Journal of Applied Crystallography. 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough, A., B. Pope, W. Chiu, and A. Weeds. 1997. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J. Cell Biol. 138:771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin, P.J., J.T. Gooch, H.G. Mannherz, and A.G. Weeds. 1993. Structure of gelsolin segment 1-actin complex and the mechanism of filament severing. Nature. 364:685–692. [DOI] [PubMed] [Google Scholar]
- Moseley, J.B., K. Okada, H.I. Balcer, D.R. Kovar, T.D. Pollard, and B.L. Goode. 2006. Twinfilin is an actin-filament-severing protein and promotes rapid turnover of actin structures in vivo. J. Cell Sci. 119:1547–1557. [DOI] [PubMed] [Google Scholar]
- Muhlrad, A., D. Kudryashov, Y. Michael Peyser, A.A. Bobkov, S.C. Almo, and E. Reisler. 2004. Cofilin induced conformational changes in F-actin expose subdomain 2 to proteolysis. J. Mol. Biol. 342:1559–1567. [DOI] [PubMed] [Google Scholar]
- Ojala, P.J., V.O. Paavilainen, M.K. Vartiainen, R. Tuma, A.G. Weeds, and P. Lappalainen. 2002. The two ADF-H domains of twinfilin play functionally distinct roles in interactions with actin monomers. Mol. Biol. Cell. 13:3811–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okreglak, V., and D.G. Drubin. 2007. Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J. Cell Biol. 178:1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otterbein, L.R., P. Graceffa, and R. Dominguez. 2001. The crystal structure of uncomplexed actin in the ADP state. Science. 293:708–711. [DOI] [PubMed] [Google Scholar]
- Paavilainen, V.O., M.C. Merckel, S. Falck, P.J. Ojala, E. Pohl, M. Wilmanns, and P. Lappalainen. 2002. Structural conservation between the actin monomer-binding sites of twinfilin and actin-depolymerizing factor (ADF)/cofilin. J. Biol. Chem. 277:43089–43095. [DOI] [PubMed] [Google Scholar]
- Paavilainen, V.O., E. Bertling, S. Falck, and P. Lappalainen. 2004. Regulation of cytoskeletal dynamics by actin-monomer-binding proteins. Trends Cell Biol. 14:386–394. [DOI] [PubMed] [Google Scholar]
- Paavilainen, V.O., M. Hellman, E. Helfer, M. Bovellan, A. Annila, M.F. Carlier, P. Permi, and P. Lappalainen. 2007. Structural basis and evolutionary origin of actin filament capping by twinfilin. Proc. Natl. Acad. Sci. USA. 104:3113–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T.D., and G.G. Borisy. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. [DOI] [PubMed] [Google Scholar]
- Quintero-Monzon, O., A.A. Rodal, B. Strokopytov, S.C. Almo, and B.L. Goode. 2005. Structural and functional dissection of the Abp1 ADFH actin-binding domain reveals versatile in vivo adapter functions. Mol. Biol. Cell. 16:3128–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rould, M.A., Q. Wan, P.B. Joel, S. Lowey, and K.M. Trybus. 2006. Crystal structures of expressed non-polymerizable monomeric actin in the ADP and ATP states. J. Biol. Chem. 281:31909–31919. [DOI] [PubMed] [Google Scholar]
- Shvetsov, A., V.E. Galkin, A. Orlova, M. Phillips, S.E. Bergeron, P.A. Rubenstein, E.H. Egelman, and E. Reisler. 2008. Actin hydrophobic loop 262-274 and filament nucleation and elongation. J. Mol. Biol. 375:793–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutt, C.E., J.C. Myslik, M.D. Rozycki, N.C. Goonesekere, and U. Lindberg. 1993. The structure of crystalline profilin-beta-actin. Nature. 365:810–816. [DOI] [PubMed] [Google Scholar]
- Tellam, R.L. 1986. Gelsolin inhibits nucleotide exchange from actin. Biochemistry. 25:5799–5804. [DOI] [PubMed] [Google Scholar]
- Witke, W. 2004. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 14:461–469. [DOI] [PubMed] [Google Scholar]
- Wriggers, W., J.X. Tang, T. Azuma, P.W. Marks, and P.A. Janmey. 1998. Cofilin and gelsolin segment-1: molecular dynamics simulation and biochemical analysis predict a similar actin binding mode. J. Mol. Biol. 282:921–932. [DOI] [PubMed] [Google Scholar]