Abstract
In physical, biological, technological and social systems, interactions between units give rise to intricate networks. These—typically non-trivial—structures, in turn, critically affect the dynamics and properties of the system. The focus of most current research on complex networks is, still, on global network properties. A caveat of this approach is that the relevance of global properties hinges on the premise that networks are homogeneous, whereas most real-world networks have a markedly modular structure. Here, we report that networks with different functions, including the Internet, metabolic, air transportation and protein interaction networks, have distinct patterns of connections among nodes with different roles, and that, as a consequence, complex networks can be classified into two distinct functional classes on the basis of their link type frequency. Importantly, we demonstrate that these structural features cannot be captured by means of often studied global properties.
The structure of complex networks1,2 is typically characterized in terms of global properties, such as the average shortest path length between nodes3, the clustering coefficient3, the assortativity4 and other measures of degree–degree correlations5,6, and, especially, the degree distribution7,8. However, these global quantities are truly informative only when one of two strict conditions is fulfilled: (1) the network lacks a modular structure9–14, or (2) the network has a modular structure but (2.1) all modules were formed according to the same mechanisms, and therefore have similar properties, and (2.2) the interface between modules is statistically similar to the bulk of the modules, except for the density of links. If neither of these two conditions is fulfilled, then any theory proposed to explain, for example, a scale-free degree distribution needs to take into account the modular structure of the network.
To our knowledge, no real-world network has been shown to fulfil either of the two conditions above; this implies that global properties may sometimes fail to provide insight into the mechanisms responsible for the formation or growth of these networks. Alternative approaches that take into consideration the modular structure of real-world complex networks are therefore necessary. One such approach is to group nodes into a small number of roles, according to their pattern of intra-and intermodule connections11–13. Recently, we demonstrated that the role of a node conveys significant information about the importance of the node, and about the evolutionary pressures acting on it11,13. Here, we demonstrate that modular networks can be classified into distinct functional classes according to the patterns of role-to-role connections, and that the definition of link types can help us understand the function and properties of a particular class of networks.
MODULARITY OF COMPLEX NETWORKS
We analyse four different types of real-world networks—metabolic networks11,15,16, protein interactomes17–20, global and regional air transportation networks13,21,22 and the Internet at the autonomous system (AS) level5,23 (Table 1 and Supplementary Information). To determine and quantify the modular structure of these networks, we use simulated annealing24 to find the optimal partition of the network into modules11,12,25 (see the Methods section). We then assess the significance of the modular structure of each network by comparing it with a randomization of the same network25. We find that all networks studied have a significant modular structure (Table 1). Modules correspond to functional units in biological networks11,20 and to geo-political units in air transportation networks13 and, probably, in the Internet26.
Table 1.
Properties and modularity of the studied networks. We show the number of nodes and links in the network, the modularity M of the best partition obtained using simulated annealing, and the average modularity (and standard deviation) of the randomizations of the network, obtained using the Markov-chain switching algorithm to preserve the degree of each node (see the Methods section). Note that all networks are significantly modular, that is, their modularity is larger than the modularity of their corresponding randomizations.
| Network type | Network | Nodes | Links | NM | M | |
|---|---|---|---|---|---|---|
| Metabolism Archaea | Archaeoglobus fulgidus | 303 | 366 | 16 | 0.813 | 0.746 (0.005) |
| Aeropyrum pernix | 300 | 387 | 14 | 0.797 | 0.711 (0.006) | |
| Methanococcus jannaschii | 223 | 277 | 14 | 0.813 | 0.720 (0.003) | |
| Pyrobaculum aerophilum | 335 | 421 | 15 | 0.811 | 0.731 (0.004) | |
| Pyrococcus furiosus | 302 | 384 | 16 | 0.813 | 0.720 (0.007) | |
| Sulfolobus solfataricus | 367 | 455 | 17 | 0.813 | 0.736 (0.006) | |
|
| ||||||
| Metabolism Bacteria | Bacillus subtilis | 649 | 863 | 20 | 0.815 | 0.724 (0.003) |
| Escherichia coli | 739 | 1,009 | 17 | 0.810 | 0.711 (0.003) | |
| Fusobacterium nucleatum | 378 | 473 | 16 | 0.816 | 0.734 (0.004) | |
| Helicobacter pylori | 360 | 438 | 15 | 0.837 | 0.746 (0.006) | |
| Mycobacterium leprae | 451 | 578 | 16 | 0.814 | 0.732 (0.005) | |
| Thermosynechococcus elongatus | 448 | 546 | 17 | 0.830 | 0.755 (0.006) | |
|
| ||||||
| Metabolism Eukaryotes | Arabidopsis thaliana | 607 | 792 | 18 | 0.825 | 0.728 (0.003) |
| Caenorhabditis elegans | 431 | 569 | 17 | 0.818 | 0.714 (0.004) | |
| Homo sapiens | 792 | 1,056 | 23 | 0.842 | 0.727 (0.003) | |
| Plasmodium falciparum | 280 | 363 | 12 | 0.815 | 0.708 (0.006) | |
| Saccharomyces cerevisiae | 570 | 776 | 17 | 0.814 | 0.708 (0.003) | |
| Schizosaccharomyces pombe | 503 | 664 | 18 | 0.827 | 0.721 (0.003) | |
|
| ||||||
| Air transportation | Global | 3,618 | 14,142 | 25 | 0.706 | 0.3111 (0.0009) |
| Asia & Middle East | 706 | 2,572 | 10 | 0.642 | 0.325 (0.002) | |
| North America | 940 | 3,446 | 12 | 0.522 | 0.3111 (0.0005) | |
|
| ||||||
| Interactome | S. cerevisiae | 1,458 | 1,948 | 25 | 0.820 | 0.707 (0.002) |
| C. elegans | 2,889 | 5,188 | 28 | 0.688 | 0.561 (0.002) | |
|
| ||||||
| Internet | 1998 | 3,216 | 5,705 | 17 | 0.625 | 0.5365 (0.0011) |
| 1999 | 4,513 | 8,374 | 18 | 0.620 | 0.5227 (0.0007) | |
| 2000 | 6,474 | 12,572 | 22 | 0.631 | 0.5042 (0.0008) | |
To assess whether global average properties are appropriate to describe the structure of these networks, we compare global average properties of the networks with the corresponding module-specific averages; specifically, we focus on the degree, the clustering coefficient and the normalized clustering coefficient. We find that the average degree of the network is not representative of individual-module average degrees for air transportation networks (Table 2). Most importantly, the global clustering coefficient is not representative of individual-module clustering coefficients for any network (except, maybe, for one out of 18 metabolic networks).
Table 2.
Global versus module-specific average properties. For each network, we show the fraction r of modules (and standard deviation) whose average degree 〈ki〉i, clustering coefficient 〈Ci 〉i, and normalized clustering coefficient significantly differ (at 95% confidence) from the global network average (see the Methods section). Fractions r > 0.05 indicate that a given global property does not correctly describe individual modules. Global degree is not representative of individual-module degrees for air transportation networks. Most importantly, the global clustering coefficient is not representative of individual-module clustering coefficients for any network (except, maybe, the metabolic network of F. nucleatum).
| Network type | Network | R〈ki〉i | r〈Ci〉i | |
|---|---|---|---|---|
| Metabolism Archaea | A. fulgidus | 0.02 (0.03) | 0.125 (0.0) | 0.10 (0.03) |
| A. pernix | 0.0 (0.0) | 0.17 (0.04) | 0.18 (0.04) | |
| M. jannaschii | 0.0 (0.0) | 0.27 (0.03) | 0.27 (0.02) | |
| P. aerophilum | 0.03 (0.03) | 0.22 (0.06) | 0.16 (0.05) | |
| P. furiosus | 0.02 (0.03) | 0.27 (0.04) | 0.24 (0.06) | |
| S. solfataricus | 0.02 (0.03) | 0.15 (0.04) | 0.11 (0.04) | |
|
| ||||
| Metabolism Bacteria | B. subtilis | 0.02 (0.02) | 0.22 (0.06) | 0.19 (0.04) |
| E. coli | 0.02 (0.04) | 0.27 (0.06) | 0.29 (0.04) | |
| F. nucleatum | 0.0 (0.0) | 0.06 (0.02) | 0.06 (0.03) | |
| H. pylori | 0.08 (0.05) | 0.28 (0.04) | 0.26 (0.03) | |
| M. leprae | 0.0 (0.0) | 0.28 (0.05) | 0.27 (0.04) | |
| T. elongatus | 0.01 (0.02) | 0.11 (0.03) | 0.12 (0.04) | |
|
| ||||
| Metabolism Eukaryotes | A. thaliana | 0.04 (0.03) | 0.29 (0.06) | 0.29 (0.07) |
| C. elegans | 0.064 (0.004) | 0.31 (0.03) | 0.30 (0.03) | |
| H. sapiens | 0.08 (0.03) | 0.45 (0.04) | 0.41 (0.05) | |
| P. falciparum | 0.084 (0.002) | 0.23 (0.03) | 0.24 (0.02) | |
| S. cerevisiae | 0.09 (0.04) | 0.24 (0.05) | 0.23 (0.05) | |
| S. pombe | 0.059 (0.003) | 0.37 (0.06) | 0.36 (0.06) | |
|
| ||||
| Air transportation | Global | 0.41 (0.05) | 0.531 (0.010) | 0.43 (0.02) |
| Asia & Middle East | 0.40 (0.10) | 0.26 (0.04) | 0.21 (0.05) | |
| North America | 0.37 (0.03) | 0.40 (0.04) | 0.47 (0.05) | |
|
| ||||
| Interactome | S. cerevisiae | 0.0 (0.0) | 0.25 (0.09) | 0.67 (0.04) |
| C. elegans | 0.042 (0.014) | 0.47 (0.06) | 0.33 (0.04) | |
|
| ||||
| Internet | 1998 | 0.064 (0.005) | 0.77 (0.05) | 0.77 (0.06) |
| 1999 | 0.0 (0.0) | 0.85 (0.03) | 0.83 (0.05) | |
| 2000 | 0.0 (0.0) | 0.77 (0.04) | 0.76 (0.07) | |
ROLE-BASED DESCRIPTION OF COMPLEX NETWORKS
As an alternative to the average description approach, we determine the role of each node according to two properties11,12 (see the Methods section): the relative within-module degree z, which quantifies how well connected a node is to other nodes in their module, and the participation coefficient P, which quantifies to what extent the node connects to different modules. We classify as non-hubs those nodes that have a low within-module degree (z < 2.5). Depending on the fraction of connections they have to other modules, non-hubs are further subdivided into11,12: (R1) ultra-peripheral nodes, that is, nodes with all their links within their own module; (R2) peripheral nodes, that is, nodes with most links within their module; (R3) satellite connectors, that is, nodes with a high fraction of their links to other modules and (R4) kinless nodes, that is, nodes with links homogeneously distributed among all modules. We classify as hubs those nodes that have a high within-module degree (z ≥ 2.5). Similar to non-hubs, hubs are divided according to their participation coefficient into: (R5) provincial hubs, that is, hubs with the vast majority of links within their module; (R6) connector hubs, that is, hubs with many links to most of the other modules and (R7) global hubs, that is, hubs with links homogeneously distributed among all modules.
Although the full rationale for this particular definition of the roles has been given elsewhere12, it is important to highlight a few properties of our classification scheme. Nodes in real and model networks, especially non-hubs, do not fill uniformly the zP-plane; our role classification scheme arises from the fact that nodes tend to congregate into a small number of densely populated regions of this space, with boundaries between these regions having a low density of nodes. In addition, especially for hubs, boundaries coincide with well-defined connectivity patterns; for example, nodes at the boundary between connector hubs (R6) and global hubs (R7) would have approximately half of their links in one module, and the other half perfectly spread in other modules. Importantly, other definitions of the roles do not alter the results we report below (see the Supplementary Information).
We investigate how our definition of roles relates to global network properties, and to what extent global network properties are representative of nodes with different roles. As some simple properties such as the degree and the clustering coefficient trivially depend on a node’s role, we focus on degree–degree correlations4–6,19,27,28. Specifically, we address two questions: (1) whether nodes with the same degree but different roles have the same or different correlations and (2) to what extent the observed degree–degree correlations are a by-product of the modular structure of the network.
To answer these questions, we start by considering the Internet at the AS level (Fig. 1). Nodes with degree k = 3 can be either ultra-peripheral (R1, if they have all connections in the same module), peripheral (R2, if they have two connections in one module and one in another) or satellite connectors (R3, if the three connections are to different modules). A separate analysis for each role reveals that the average degree knn(k) of the neighbours of a node5 with degree k = 3 strongly depends on the role of the node. For an instance of the 1998 Internet, for example, knn(k = 3) = 43 ± 8 for ultra-peripheral nodes, knn(k = 3) = 196 ± 12 for peripheral nodes and knn(k = 3) = 290 ± 20 for satellite connectors. We observe a dependence of knn on the nodes’ role for all the networks studied here (Fig. 1a–d).
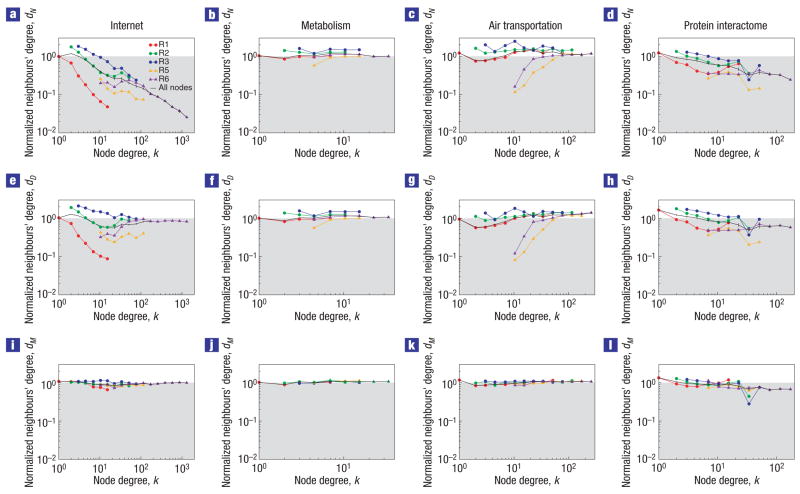
Figure 1. Modularity and degree distribution explain most degree–degree correlations in complex networks.
a–d, Degree dN of the neighbours of a node normalized by the average neighbours’ degree of all the nodes in the network. e–h, Degree dD of the neighbours of a node normalized by the average neighbours’ degree of the node in the ensemble of random networks with fixed degree sequence. i–l, Neighbours’ degree dM of a node normalized by the average neighbours’ degree of the node in the ensemble of random networks with fixed degree sequence and modular structure (see the Methods section). Values of d are averaged over nodes with similar degrees to obtain the function d(k). The error bars represent the standard error of the average. Note that a lack of deviations from the ensemble average, that is, d(k) = 1, indicates the absence of correlations. The results in the middle row show that the degree distribution is responsible for some of the observed degree–degree correlations, but cannot fully account for them. The degree distribution and the modular structure of the network do account for most existing degree–degree correlations in the Internet, metabolic and air transportation networks.
Regarding the second question, initial research showed5 that for the Internet at the AS level knn(k) ∝ k−0.5. It was later pointed out27,28 that any network with the same degree distribution as the Internet should exhibit a similar scaling. In other words, the degree distribution of the network is responsible for most of the observed correlations. However, the degree distribution alone does not account for all the observed correlations28 (Fig. 1e). In contrast, the modular structure of the network does account for most of the remaining degree–degree correlations observed in the topology of the Internet (Fig. 1i). Similarly, the modular structure accounts for the degree–degree correlations in metabolic networks and the air transportation network, and for most of the correlations in protein interaction networks (Fig. 1i–l).
ROLE-TO-ROLE CONNECTIVITY PROFILES
The findings we reported so far suggest that, once the degree distribution and the modular structure are fixed, real networks have no additional internal structure. This, however, contradicts our intuition that networks with different growth mechanisms and functional needs should have distinct connection patterns between nodes playing different roles. To investigate this possibility, we systematically analyse how nodes connect to one another depending on their roles.
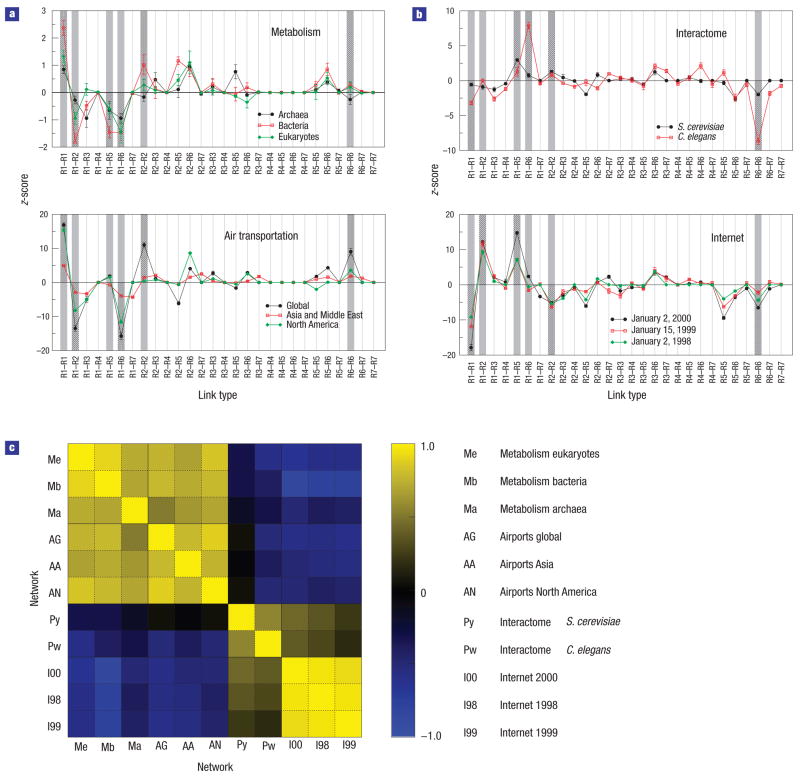
For each network, we calculate the number rij of links between nodes belonging to roles i and j, and compare this number to the number of such links in a properly randomized network (see the Methods section). As in previous work19,28–30, we use the z-score to obtain a profile a of over- and under-representation of link types (Fig. 2), which enables us to compare different networks. We quantify the overall similarity between two profiles, a and b, by the scalar product between these profiles (see the Methods section). In Fig. 2, we show that networks of the same type have highly correlated profiles, whereas networks of different types have weaker correlations and, at times, even strong anti-correlations (Fig. 2c).
Figure 2. Role-to-role connectivity patterns.
a,b, The z-score for the abundance (see the Methods section) of each link type for stringy-periphery networks (a) and multi-star networks (b), see text. Roles are labelled as follows: (R1) ultra-peripheral; (R2) peripheral; (R3) satellite connectors; (R4) kinless nodes; (R5) provincial hubs; (R6) connector hubs; (R7) global hubs. c, Quantification of the similarity between two z-score profiles by means of the correlation coefficient (see the Methods section), with yellow corresponding to large positive correlation, blue to large anti-correlation and black to no correlation. The grey columns in a indicate those link types that contribute the most, in absolute value, to the correlation coefficient. These link types are, therefore, the ones that better characterize the set of all profiles.
The networks considered fall into two main classes, one comprising metabolic and air transportation networks, and another comprising protein interactomes and the Internet. The main difference between the two groups is the pattern of links between: (1) ultra-peripheral nodes (links of type R1–R1) and (2) connector hubs and other hubs (links of types R5–R6 and R6–R6). These link types are over-represented for networks in the first class (except links of type R6–R6 in metabolic networks), and under-represented for networks in the second class.
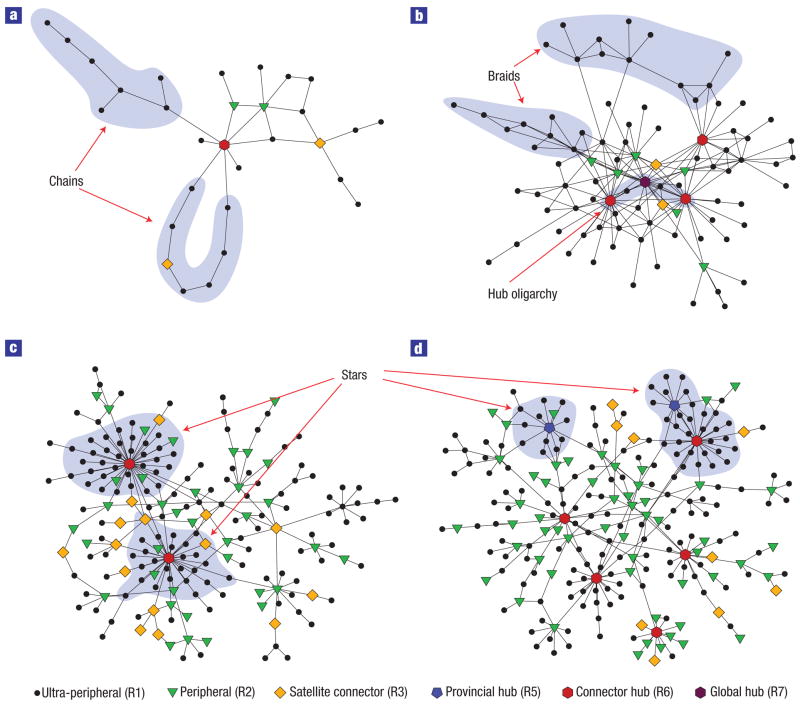
We denote the first class as the stringy-periphery class (Fig. 3a,b). In networks of this class, ultra-peripheral nodes are more connected to one another than would be expected from chance, which results in long ‘chains’ of ultra-peripheral nodes. In metabolic networks, these chains correspond to loop-less pathways that, for example, degrade a complex metabolite into simpler molecules. In the air transportation network, owing to the higher overall connectivity of the network, chains contain short loops and resemble ‘braids’. Stringy-periphery networks also have a core of hubs, which we call the hub oligarchy, that are directly reachable from one another (links of type R5–R6 in metabolic and air transportation networks, and R6–R6 in air transportation networks). Moreover, connector hubs are less connected to ultra-peripheral nodes (R1) than expected by chance alone.
Figure 3. Modules and role-to-role connectivity signatures in different network types.
a–d, Representation of a single module (that is, all the nodes depicted belong to a single module) in the metabolic network of A. thaliana (a), the Asia and Middle East air transportation network (b), the protein interactome of C. elegans (c) and the Internet in 1998 (d). Different symbols indicate different node roles (see Supplementary Information for the names of the nodes). External links to other modules are not depicted, although it is possible to infer where they are from the role of the nodes. The shaded regions highlight important structural features.
We denote the second class as the multi-star class (Fig. 3c,d). The multi-star class comprises the protein interactomes and the Internet, and has the opposite signature to the stringy-periphery class. Links of type R1–R1 (between ultra-peripheral nodes) are under-represented, whereas links of type R1–R5 (between ultra-peripheral nodes and provincial hubs) are over-represented, giving rise to modules with indirectly connected ‘star-like’ structures. Similarly, connector hubs are less connected to one another than would be expected, which means that these networks depend on satellite connectors to bridge connector hubs and modules.
Our findings confirm and clarify previous results in the literature. For example, the under-representation of R6–R6 links in protein interactomes is consistent with previous results suggesting a tendency for hubs to ‘repel’ each other in these networks6,19. Similarly, the role-to-role connectivity profile of the Internet is consistent with the existence of a hierarchy of types of nodes28. This hierarchy comprises end users, regional providers and global providers, which we hypothesize correspond to roles R1–R2, R5 and R6 respectively. The role-to-role connectivity profiles are consistent with a scenario in which end users connect mostly to regional providers, and in which global providers connect with each other indirectly through satellite connectors (R3), with few connections but probably large bandwidth.
By considering the modular structure of the networks and the extra dimension introduced by the participation coefficient, however, our approach provides novel insights into the relationship between structure and function in complex networks. For example, by considering the absolute degree alone, nodes with roles R5 and R6 in protein interactomes are indistinguishable from each other: in Saccharomyces cerevisiae, 〈k〉R5 = 14.0 ± 1.7 and 〈k〉R6 = 17.1 ± 1.9, whereas the average degree for the whole network is 〈k〉 = 2.67±0.09. Still, links R5–R5 between provincial hubs, unlike R6–R6 links, are not under-represented. In general, the different connection patterns of R5 and R6 (or R1 and R2) proteins enables us to hypothesize that they play distinct biological roles, with R6 proteins probably being much more important31.
A closer look at the air transportation network also helps to show that important structural properties may be left unexplained by focusing on degree alone, as well as to stress the importance of the relative within-module degree as opposed to the degree. Johannesburg, in South Africa, has degree k = 84, which is 23% smaller than the degree of Cincinnati in the US, k = 109. Still, it is possible to fly from most capitals in the world to Johannesburg but not to Cincinnati. There are two main reasons for this. First, although Johannesburg is the most connected city in its region (sub-Saharan Africa), Cincinnati (North America) is not; this effect is captured by the within-module relative degree, which is 9.3 for Johannesburg and 4.3 for Cincinnati. Second, Johannesburg has many connections to other regions, whereas Cincinnati does not; this effect is captured by the participation coefficient, which is 0.52 for Johannesburg and 0.05 for Cincinnati. As a result, Johannesburg is a global hub (R6) in our classification, whereas Cincinnati is a provincial hub (R5). Thus, it can be understood why R6–R6 connections are over-represented in air transportation networks (most global hubs are connected to one another), whereas R5–R5 are not (most provincial hubs are poorly connected to provincial hubs in other regions). In general, our approach shows why the behaviour of R5 and R6 nodes is so different in air transportation networks, which cannot be understood from the degree of the nodes alone.
We have shown that global properties that do not take into account the modular organization of the network may sometimes fail to capture potentially important structural features; although all networks (except, maybe, the protein interactomes) show no degree–degree correlations when compared with the appropriate ensemble of random networks, they all have clearly distinctive properties in terms of how nodes with certain roles are connected to each other. Our results thus call attention to the need to develop new approaches that will enable us to better understand the structure and evolution of real-world complex networks.
In addition, our findings demonstrate that networks with the same functional needs and growth mechanisms have similar patterns of connections between nodes with different roles. Attempts to divide complex networks into ‘classes’ or ‘families’ have been made before, for example, in terms of the degree distribution8 and in terms of the relative abundance of certain subgraphs or motifs29,30. Our work here complements those attempts, and is the first one to build on the crucial fact that most real-world networks exhibit a markedly modular structure.
Although we cannot put forward a theory for the division of the networks into two classes, we hypothesize that it might be related to the fact that networks in the stringy-periphery class are transportation networks, in which strict conservation laws must be fulfilled. Indeed, for transportation systems it has been shown that, under quite general conditions, a hub oligarchy is the the most efficient organization32. Conversely, both protein interactomes and the Internet can be seen as signalling networks, which do not obey conservation laws.
METHODS
MODULE IDENTIFICATION
The modularity ℳ (℘) of a partition ℘ of a network into modules is10
where NM is the number of non-empty modules (smaller than or equal to the number N of nodes in the network), L is the number of links in the network, ls is the number of links between nodes in module s and ds is the sum of the degrees of the nodes in module s. The objective of a module identification algorithm is to find the partition ℘* that yields the largest modularity M≡ℳ(℘*). Note that NM is only constrained to be NM ≤ N, but is otherwise selected by the optimization algorithm so that M is maximum. The problem of identifying the optimal partition is analogous to finding the ground state of a disordered system with hamiltonian ℋ=−Lℳ (ref. 25).
As the modularity landscape is in general very rugged, we use simulated annealing to find a close to optimal partition of the network into modules11,12,25. This method is the most accurate to date11,14.
ROLE DEFINITION
We determine the role of each node according to two properties11,12: the relative within-module degree z and the participation coefficient P. The within-module degree z-score measures how ‘well-connected’ node i is to other nodes in the module compared with those other nodes, and is defined as
where is the number of links of node i to nodes in module s, si is the module to which node i belongs, and the averages 〈…〉j∈s are taken over all nodes in module s.
The participation coefficient quantifies to what extent a node connects to different modules. We define the participation coefficient Pi of node i as
Where is the number of links of node i to nodes in module s, and is the total degree of node i. The participation coefficient of a node is therefore close to one if its links are uniformly distributed among all the modules and zero if all its links are within its own module.
We classify as non-hubs those nodes that have a low within-module degree (z < 2.5). Depending on the amount of connections they have to other modules, non-hubs are further subdivided into11,12: (R1) ultra-peripheral nodes, that is, nodes with all their links within their own module (P ≤ 0.05); (R2) peripheral nodes, that is, nodes with most links within their module (0.05 < P ≤ 0.62); (R3) satellite connectors, that is, nodes with a high fraction of their links to other modules (0.62 < P ≤ 0.80) and (R4) kinless nodes, that is, nodes with links homogeneously distributed among all modules (P > 0.80). We classify as hubs those nodes that have a high within-module degree (z ≥ 2.5). Similar to non-hubs, hubs are divided according to their participation coefficient into: (R5) provincial hubs, that is, hubs with the vast majority of links within their module (P ≤ 0.30); (R6) connector hubs, that is, hubs with many links to most of the other modules (0.30 < P ≤ 0.75) and (R7) global hubs, that is, hubs with links homogeneously distributed among all modules (P > 0.75).
NETWORK RANDOMIZATION AND STATISTICAL ENSEMBLES
We use two different ensembles of random networks19,28. In the first ensemble, which we denote by
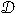 , we only preserve the degree sequence of the original network; in the second ensemble, denoted ℳ, we preserve both the degree sequence and the modular structure of the network. Averages over the first and second ensembles are denoted 〈⋯〉
, we only preserve the degree sequence of the original network; in the second ensemble, denoted ℳ, we preserve both the degree sequence and the modular structure of the network. Averages over the first and second ensembles are denoted 〈⋯〉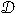 and 〈⋯〉ℳ, respectively.
and 〈⋯〉ℳ, respectively.
To generate random networks in ensemble
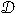 , we randomize all the links in the network while preserving the degree of each node. To uniformly sample all possible networks, we use the Markov-chain Monte Carlo switching algorithm19,33. In this algorithm, we repeatedly select random pairs of links, for example (i, j) and (l, m), and swap one of the ends of each link, so that the links become (i, m) and (l, j).
, we randomize all the links in the network while preserving the degree of each node. To uniformly sample all possible networks, we use the Markov-chain Monte Carlo switching algorithm19,33. In this algorithm, we repeatedly select random pairs of links, for example (i, j) and (l, m), and swap one of the ends of each link, so that the links become (i, m) and (l, j).
To generate random networks in ensemble ℳ, we restrict the Markov-chain Monte Carlo switching algorithm28 to pairs of links that connect nodes in the same pair of modules, that is, we apply the Markov-chain Monte Carlo switching algorithm independently to links whose ends are in modules 1 and 1, 1 and 2, and so forth for all pairs of modules. This method guarantees that, with the same partition as the original network, the modularity of the randomized network is the same as that of the original network (as the number of links between each pair of modules is unchanged) and that the role of each node is also preserved.
To investigate whether global properties are representative of module-specific properties, we focus on degree ki, clustering coefficient Ci and normalized clustering coefficient For each module s in the network, comprising ns nodes, we compute the average of each property in the module (for example, 〈ki 〉i∈s). In addition, we compute the distribution of such averages for random modules, which we obtain by randomly selecting groups of ns nodes. If the empirical module average falls outside of the 95% probability of the distribution for the random modules, we consider that the global average is not representative of the module average. We finally compute the fraction r of modules that are not properly described by the global average.
To study degree–degree correlations, we consider the average degree of the nearest neighbours of each node i. We define the normalized nearest-neighbours’ degree di as the ratio of and: (1) the average value of in the network
where N is the number of nodes in the network; (2) the expected value of in the ensemble of networks with fixed degree sequence
and (3) the expected value of in the ensemble of networks with fixed degree sequence and modular structure
Note that, in spite of the similar notation, the meaning of is somewhat different from the other two because the normalization involves an average over nodes, whereas in and the normalization involves averages over an ensemble of randomized networks.
To obtain the role-to-role connectivity profiles, we calculate the z-score19,28–30 of the number of links between nodes with roles i and j as
where rij is the number of links between nodes with roles i and j. To obtain better statistics and an estimation of the error in the z-score, we carry out this process for several partitions of each network.
To evaluate the similarity between two z-score profiles, a and b, we use the scalar product
where σza is the standard deviation of the elements in a.
Acknowledgments
We thank R. D. Malmgren, E. N. Sawardecker, S. M. D. Seaver, D. B. Stouffer and M. J. Stringer for useful comments and suggestions. R.G. and M.S.-P. thank the Fulbright Program. L.A.N.A. gratefully acknowledges the support of a NIH/NIGMS K-25 award, of NSF award SBE 0624318, of the J. S. McDonnell Foundation and of the W. M. Keck Foundation.
Footnotes
Competing financial interests
The authors declare that they have no competing financial interests.
Reprints and permission information is available online at http://npg.nature.com/reprintsandpermissions/
Supplementary Information accompanies this paper on www.nature.com/naturephysics.
References
- 1.Newman MEJ. The structure and function of complex networks. SIAM Rev. 2003;45:167–256. [Google Scholar]
- 2.Amaral LAN, Ottino J. Complex networks: Augmenting the framework for the study of complex systems. Eur Phys J B. 2004;38:147–162. [Google Scholar]
- 3.Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 4.Newman MEJ. Assortative mixing in networks. Phys Rev Lett. 2002;89:208701. doi: 10.1103/PhysRevLett.89.208701. [DOI] [PubMed] [Google Scholar]
- 5.Pastor-Satorras R, Vázquez A, Vespignani A. Dynamical and correlation properties of the Internet. Phys Rev Lett. 2001;87:258701. doi: 10.1103/PhysRevLett.87.258701. [DOI] [PubMed] [Google Scholar]
- 6.Colizza V, Flammini A, Serrano MA, Vespignani A. Detecting rich-club ordering in complex networks. Nature Phys. 2006;2:110–115. [Google Scholar]
- 7.Barabási AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 8.Amaral LAN, Scala A, Barthélémy M, Stanley HE. Classes of small-world networks. Proc Natl Acad Sci USA. 2000;97:11149–11152. doi: 10.1073/pnas.200327197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Girvan M, Newman MEJ. Community structure in social and biological networks. Proc Natl Acad Sci USA. 2002;99:7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newman MEJ, Girvan M. Finding and evaluating community structure in networks. Phys Rev E. 2004;69:026113. doi: 10.1103/PhysRevE.69.026113. [DOI] [PubMed] [Google Scholar]
- 11.Guimerà R, Amaral LAN. Functional cartography of complex metabolic networks. Nature. 2005;433:895–900. doi: 10.1038/nature03288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guimerà R, Amaral LAN. Cartography of complex networks: modules and universal roles. J Stat Mech Theor Exp. 2005:P02001. doi: 10.1088/1742-5468/2005/02/P02001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guimerà R, Mossa S, Turtschi A, Amaral LAN. The worldwide air transportation network: Anomalous centrality, community structure, and cities’ global roles. Proc Natl Acad Sci USA. 2005;102:7794–7799. doi: 10.1073/pnas.0407994102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danon L, Díaz-Guilera A, Duch J, Arenas A. Comparing community structure identification. J Stat Mech Theor Exp. 2005:P09008. [Google Scholar]
- 15.Jeong H, Tombor B, Albert R, Oltvai ZN, Barabási AL. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 16.Wagner A, Fell DA. The small world inside large metabolic networks. Proc R Soc B. 2001;268:1803–1810. doi: 10.1098/rspb.2001.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uetz P, et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 18.Jeong H, Mason SP, Barabási AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 19.Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296:910–913. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- 20.Li S, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrat A, Barthélemy M, Pastor-Satorras R, Vespignani A. The architecture of complex weighted networks. Proc Natl Acad Sci USA. 2004;101:3747–3752. doi: 10.1073/pnas.0400087101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, Cai X. Statistical analysis of airport network of China. Phys Rev E. 2004;69:046106. doi: 10.1103/PhysRevE.69.046106. [DOI] [PubMed] [Google Scholar]
- 23.Vázquez A, Pastor-Satorras R, Vespignani A. Large-scale topological and dynamical properties of the Internet. Phys Rev E. 2002;65:066130. doi: 10.1103/PhysRevE.65.066130. [DOI] [PubMed] [Google Scholar]
- 24.Kirkpatrick S, Gelatt CD, Vecchi MP. Optimization by simulated annealing. Science. 1983;220:671–680. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- 25.Guimerà R, Sales-Pardo M, Amaral LAN. Modularity from fluctuations in random graphs and complex networks. Phys Rev E. 2004;70:025101. doi: 10.1103/PhysRevE.70.025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eriksen KA, Simonsen I, Maslov S, Sneppen K. Modularity and extreme edges of the Internet. Phys Rev Lett. 2003;90:148701. doi: 10.1103/PhysRevLett.90.148701. [DOI] [PubMed] [Google Scholar]
- 27.Park J, Newman MEJ. Origin of degree correlations in the Internet and other networks. Phys Rev E. 2003;68:026112. doi: 10.1103/PhysRevE.68.026112. [DOI] [PubMed] [Google Scholar]
- 28.Maslov S, Sneppen K, Zaliznyak A. Detection of topological patterns in complex networks: correlation profile of the internet. Physica A. 2004;333:529–540. [Google Scholar]
- 29.Milo R, et al. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 30.Milo R, et al. Superfamilies of evolved and designed networks. Science. 2004;303:1538–1542. doi: 10.1126/science.1089167. [DOI] [PubMed] [Google Scholar]
- 31.Han JDJ, et al. Evidence for dynamically organized modularity in the yeast protein–protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 32.Arenas A, Cabrales A, Díaz-Guilera A, Guimerà R, Vega-Redondo F. Statistical Mechanics of Complex Networks. In: Pastor-Satorras R, Rubi M, Díaz-Guilera A, editors. Lecture Notes in Physics. Ch 10. Springer; Berlin: 2003. pp. 175–194. [Google Scholar]
- 33.Itzkovitz S, Milo R, Kashtan N, Newman MEJ, Alon U. Reply to Comment on ‘Subgraphs in random networks’. Phys Rev E. 2004;70:058102. doi: 10.1103/PhysRevE.68.026127. [DOI] [PubMed] [Google Scholar]