Whenever information is transmitted, there is an inherent risk of errors and, concurrently, the question of how to correct those errors. One approach is to introduce redundancy in the transmitted message, for example, by adding a parity bit to a transmitted number. In this issue of PNAS, Tsvi Tlusty (1) studies a similar problem in the context of biology, in particular, of molecular codes. The importance of this work lies in the generality of the questions it asks and the elegant solutions it provides.
The basic question can best be understood in terms of a key example: the genetic code (ref. 2, and see also ref. 3). In this case, the problem involves associating an amino acid with any triplet (“word”) of nucleotides (usually represented by the four letters A, C, G, and T). Because the words representing the amino acids are made up of three letters, each of which has four possible values, one could, in principle, code for 64 = 43 different outcomes. Therefore, given its structure, the genetic code can, in principle, code for 64 different amino acids.
However, nature codes for only 20 amino acids, and one feels that this limitation should somehow compensate for the intrinsic molecular noise that is present in any biological process and that can lead to faulty readout of some of the letters. The merit of Tlusty's work (1) is that it proposes a general method of formulating problems of this kind and argues convincingly for the case described above: that 20 is about the “right” number, in terms of both evolutionary constraints and noise. This insight is based on a number of mathematical constructions. In particular, Tlusty's work combines rate distortion theory (4) and statistical mechanics. In this commentary, I do not profess to explain the details of the construction but will try to convey a picture of what is involved.
The basic idea is that the genetic code (or, as in ref. 1, some molecular code) must cope with two conflicting issues: coding many outcomes (the number 20, above) while minimizing readout errors. In short, a totally error-free code would be one that codes “nothing,” that is, all triplets of letters A, C, G, and T lead to only one amino acid, so readout errors do not matter. This is very precise but not very useful. At the other extreme, one could code for 64 amino acids, but then any readout error will lead to assigning the wrong amino acid. In biological systems, there is a third issue, taken into account in ref. 1, which is the cost of actually constructing the decoder. This adds another layer of optimization that I will not discuss further here.
The essential inputs to the problem are the probability of making a readout error and the average impact of such a mistake, which is called the “error load.” For example, in the case of the genetic code, the probability of misreading one of the three letters is much higher than that of misreading two or even all three. Therefore, it is essential that the code minimize the impact of one-letter misreading by assigning, if possible, the same amino acid to codons that differ by only a single letter. Tlusty assumes in his work (1) that the reading of T and C is always confused in the last position of the word (5), and in this way he arrives at only 48 possible codons (4·4·3).
Given these error probabilities, one can now ask what the “optimal” number of amino acids might be. Tlusty (1) formulates this problem as follows. The 48 codons are viewed as nodes on a graph, as shown in Fig. 1. Two nodes are linked if their codons differ in exactly one letter. (There are, of course, numerous other representations, some more appealing than Fig. 1, but this one has the merit of mapping the error load exactly.) Say that we want to put 20 different “colors” (amino acids) on this graph. It is intuitively clear that it is better to place all codons coding for amino acid 1 (e.g., leucine) as compactly as possible, in the sense of the connectivity of the graph. Because misreading of a given codon in one letter leads to a hop on the graph across one link, if all the nodes coding for the same amino acid are close together, then the probability of hopping to coding for another amino acid will be lower if more neighboring codons (where only one of the three letters has changed) code for the same amino acid. My simplified view is that one wants to minimize the “surface” of the region coding the same amino acid relative to its “volume,” where surface and volume are counted in numbers of links and nodes in the graph.
Fig. 1.

The codon graph. Each node of the graph represents 1 of the 48 distinguishable codons. Two nodes are linked if the corresponding codons differ in exactly one position, e.g., TTG and TTA. The amino acids obtained from the codons are color-coded, with the most prominent being leucine (red) and arginine (blue). The colored links denote reading errors that do not affect the result, whereas the transparent links show reading errors that would affect the result. The optimization problem consists in maximizing the ratio of colored links over noncolored ones while also maximizing the number of different colors. Note that distances are not important in the graph; what matters is only whether two nodes are connected.
Rather than thinking in terms of graphs, imagine a chessboard (Fig. 2). It has 64 squares (here, two “letters” form the code: a to h for the files and 1 to 8 for the ranks). If we want to code for 32 black and 32 white, it is certainly much better to color half the board in black and the other half in white, rather than to use checkerboard coloring. In the first case, the probability of miscoding is 1/8 per reading error, whereas in the second it is 1 per reading error (because the color changes after any horizontal or vertical step).
Fig. 2.
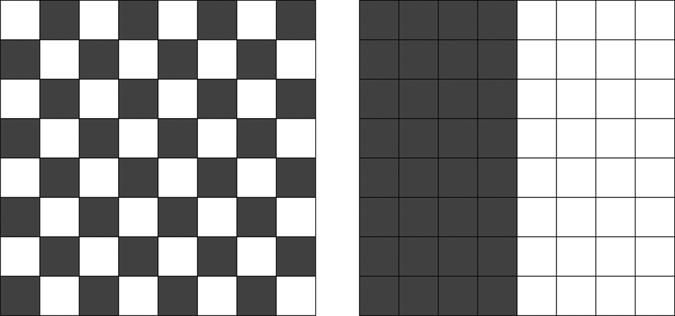
The chessboard example. Both chessboards code for two colors: black and white. A reading error occurs if one moves from one square to the horizontally or vertically adjacent square; a coding error is mistaking black for white (or vice versa). Clearly, the configuration on the left is much more likely to result in error than the one on the right.
Tlusty (1) maps the problem of finding the optimal code onto a problem of statistical mechanics, considering the boundaries of the connected regions as polymers and the error cost as an interaction on their boundary. Using the classical theory of phase transitions and polymers, he obtains the interesting result that a first-order phase transition occurs as the number of coded entities changes. One can view this as a relatively sharp increase in the expected error as a function of how many things one wants to code, all other parameters being held fixed. In other words, coming back to the example of the genetic code, the error rate shows a sharp increase once one wants to go beyond ≈20 amino acids (see figure 3B in ref. 1).
Tlusty maps the problem of finding the optimal code onto a problem of statistical mechanics.1).
This commentary has certainly glossed over a large number of relevant details that the reader can only fully understand by actually reading the article (1) (and also refs. 2 and 6); however, I hope I have given the gist of the argument. What I like in Tlusty's approach is the combination of geometry, probability theory, and analysis. In my view, this combination opens a new window to understanding the “why” of certain biological facts, which supplements beautifully the many detailed “how's” offered by current biological research. The generality of the method should also apply to other fields of research where “coding” is involved, such as language, or perhaps music. For language, the question is how, given the ambiguity of words, a message can be transmitted in a noisy environment in such a way that the receiver of the message understands what is being said, that is, the meaning of the text. In music, researchers (7, 8) have shown the geometric structure of chords and sequences. An interesting challenge would be to identify what these elements code in terms of mood, the color of the sound, and the like, that is, to connect the geometry with the meaning.
Acknowledgments.
This work was supported in part by the Fonds National Suisse.
Footnotes
The author declares no conflict of interest.
See companion article on page 8238.
References
- 1.Tlusty T. Casting polymer nets to optimize noisy molecular codes. Proc Natl Acad Sci USA. 2008;105:8238–8243. doi: 10.1073/pnas.0710274105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tlusty T. A model for the emergence of the genetic code as a transition in a noisy information channel. J Theor Biol. 2007;249:331–342. doi: 10.1016/j.jtbi.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 3.Parker G, Smith J. Optimality theory in evolutionary biology. Nature. 1990;348:27–33. [Google Scholar]
- 4.Berger T. Rate Distortion Theory: A Mathematical Basis for Data Compression. Englewood Cliffs, NJ: Prentice–Hall; 1971. Prentice–Hall Series in Information and System Sciences. [Google Scholar]
- 5.Crick FH. Codon–anticodon pairing: The wobble hypothesis. J Mol Biol. 1966;19:548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- 6.Tlusty T. A relation between the multiplicity of the second eigenvalue of a graph Laplacian, Courant's nodal line theorem and the substantial dimension of tight polyhedral surfaces. Electron J Linear Algebra. 2007;16:315–324. [Google Scholar]
- 7.Hall RW. Geometrical music theory. Science. 2008;320:328–329. doi: 10.1126/science.1155463. [DOI] [PubMed] [Google Scholar]
- 8.Callendar C, Quinn I, Tymoczko D. Generalized voice-leading spaces. Science. 2008;320:346–348. doi: 10.1126/science.1153021. [DOI] [PubMed] [Google Scholar]


