Abstract
The cytochrome P450 enzyme CYP119, its compound II derivative, and its nitrosyl complex were studied by iron K-edge x-ray absorption spectroscopy. The compound II derivative was prepared by reaction of the resting enzyme with peroxynitrite and had a lifetime of ≈10 s at 23°C. The CYP119 nitrosyl complex was prepared by reaction of the enzyme with nitrogen monoxide gas or with a nitrosyl donor and was stable at 23°C for hours. Samples of CYP119 and its derivatives were studied by x-ray absorption spectroscopy at temperatures below 140 (K) at the Advanced Photon Source of Argonne National Laboratory. The x-ray absorption near-edge structure spectra displayed shifts in edge and pre-edge energies consistent with increasing effective positive charge on iron in the series native CYP119 < CYP119 nitrosyl complex < CYP119 compound II derivative. Extended x-ray absorption fine structure spectra were simulated with good fits for k = 12 Å−1 for native CYP119 and k = 13 Å−1 for both the nitrosyl complex and the compound II derivative. The important structural features for the compound II derivative were an iron-oxygen bond length of 1.82 Å and an iron-sulfur bond length of 2.24 Å, both of which indicate an iron-oxygen single bond in a ferryl-hydroxide, FeIVOH, moiety.
Keywords: EXAFS, ferryl-oxo, XANES
The ubiquitous cytochrome P450 enzymes (P450s) are catalysts for a vast number of oxidation reactions in nature (1). They are medicinally important because they are linked to a number of disease states in man, including liver disease and breast cancer, and pharmacologically important because hepatic P450s oxidize drugs, pro-drugs, and other xenobiotics. The P450s are heme-containing proteins with a cysteine thiolate as the fifth ligand to the heme iron. The reactive oxidizing intermediates in P450s are commonly assumed to be similar to the so-called compound I complexes in other heme-containing enzymes. Compound I species are iron(IV)-oxo porphyrin radical cations, which are formed by reaction of peroxidase and catalase enzymes with hydrogen peroxide (2). Unlike other heme-containing enzymes, P450s in nature are activated by a series of rapid processes that occur after substrate binding, reduction of iron from +3 to +2 oxidation state, reversible binding of oxygen, a second reduction, and two protonation reactions (1, 3). High valent iron-oxo species in P450 enzymes are extremely short-lived and could not be observed under cryogenic reduction conditions that permitted detection of the immediate precursor to the oxidant(s), a hydroperoxy-iron(III) species (4, 5). In fact, no P450 oxidant has been detected under natural conditions, although spectroscopic evidence exists for production of iron-oxo species in low conversion under some unnatural conditions (6–9), and the identities of the P450 oxidants remain in doubt (10, 11).
The difficulty in producing P450 oxidants led to the development of a photo-oxidation method for formation of compound I analogues (12). For a model compound and horseradish peroxidase enzyme, photoejection of an electron from compound II derivatives, iron(IV)-oxo neutral porphyrin species, gave the known compound I species (12), and this method was extended to studies of a cytochrome P450 enzyme (9). In the P450 study, the putative compound II species from cytochrome P450 119 (CYP119) was formed by reaction of the resting enzyme with peroxynitrite (PN), a reaction analogous to those previously reported for other heme-containing enzymes (13) including another P450 enzyme, P450 BM3 (or CYP102) (14), and the heme-thiolate protein chloroperoxidase (CPO) (9, 14, 15). Recently, however, Green and coworkers (16) reported that reaction of CYP102 with PN gave a nitrosyl species instead of a compound II derivative.
Compound II derivatives of P450 enzymes (P450-II) are inherently interesting species in their own right, and they also are essentially uncharacterized. The interest arises mainly because P450-II is related to the transients or transition structures that are formed in the course of a carbon-hydrogen bond hydroxylation reaction catalyzed by a P450 enzyme. Unfortunately, the only evidence for and characterization of P450-II species are the UV-visible spectra of the products from reactions of CYP102 and CYP119 with PN (9, 14) and the photochemistry of the thus produced CYP119 derivative (9). For most heme enzymes, the compound II species are thought to be conventional ferryl-oxo species containing iron-oxygen double bonds, but Green and coworkers (17, 18) reported that the compound II derivative of CPO has a long iron-oxygen bond reflecting an Fe-OH moiety; i.e., a basic ferryl group that is protonated at physiological pH. More recently, however, the same group claimed that the CPO-II preparation actually gave a mixture of ferryl-oxo and ferryl hydroxide products (19). Analogues of P450 compound II species formed by chemical oxidations of the enzymes followed by internal redox reactions also apparently have basic ferryl groups as deduced from Mössbauer spectroscopy studies (20).
In this work, we report characterization of the compound II derivative from CYP119 (CYP119-II) using x-ray absorption spectroscopy (XAS). The CYP119-II species was formed by oxidation of the enzyme with PN as before (9). This transient was demonstrated to be distinct from the nitrosyl complex by comparison of the XAS spectra with that of authentic nitrosyl complex and by kinetic studies of the decomposition of CYP119-II under conditions where the nitrosyl complex is stable. An important finding from the XAS studies is that the Fe-O bond in CYP119-II is relatively long, consistent with an iron-oxygen single bond in an Fe-OH moiety, which indicates a high O-H bond strength due to the trans thiolate ligand on iron.
Results and Discussion
CYP119 is a P450 enzyme from the thermophile Sulfolobus solfataricus (7, 21–23). The enzyme used in this work was expressed in Escherichia coli and was of high purity as judged from the ratio of the absorbance at λ = 415 nm (λmax for Soret band of the heme group) to that at 280 nm, the R/Z ratio. Specifically, all samples had R/Z > 1.4.
Samples of the CYP119-II derivative were prepared by reaction of the resting enzyme with solutions of PN (24). Because the product(s) from PN reactions with P450 enzymes were questioned, we revisited this reaction. PN is prepared and stored in basic solutions (24). It is highly reactive with many biological substrates (25), and it rapidly decays in neutral solutions through an acid-catalyzed process giving approximately equal amounts of nitrate and nitrite (26–28). In addition, iron-containing enzymes catalyze the decomposition of PN (13, 25).
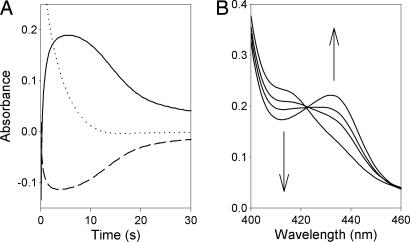
Stopped-flow mixing experiments of CYP119 with PN gave kinetics results such as those shown in Fig. 1A. The initial rate of decay of native CYP119 (λmax = 417 nm) matches that of formation of CYP119-II (λmax = 433 nm) with the conversion complete within 2 s. The CYP119-II derivative was persistent when PN was present and began to decay after the PN was depleted. The CYP119-II decay was nonexponential, but an approximate first-order rate constant for decay at ambient temperature was estimated to be k ∼ 0.15 s−1 from traces observed after decay of ≈50% of the CYP119-II species. The product from decay of CYP119-II is spectroscopically indistinguishable from resting enzyme, but previous studies of reactions of heme-containing enzymes with PN indicate that the P450 enzyme could contain nitrated amino acid residues (13, 14). The room-temperature lifetime of CYP119-II in pH 7.4 mixtures produced from reactions with excess PN typically was ≈10 s, which is more than adequate time to ensure that reaction mixtures prepared at 0°C contained CYP119-II when they were rapidly cooled to 77 K.
Fig. 1.
Kinetic results. (A) Stopped-flow kinetic traces for reaction of 15 μM CYP119 with 0.9 mM peroxynitrite at pH 7.4 and 23°C; CYP119-II was monitored at 433 nm (solid line), native CYP119 was monitored at 417 nm (dashed line), and peroxynitrite was monitored at 302 nm (dotted line). (B) Spectra from reaction of ferric CYP119 (5 μM) with sodium nitrite (0.3 M) in 50 mM phosphate buffer (pH 7.4) at 23°C; the spectra were recorded at 1.4, 20, 30, and 100 s after mixing, and the rates of signal growth at 433 nm and decay at 417 nm are equal with a pseudo-first-order rate constant of k = 0.028 s−1.
For the experiment shown in Fig. 1A, PN decay was rapid with a half-life of ≈2 s. Under otherwise similar conditions but without CYP119, the half-life of the PN was 4 s. Similar catalysis behavior for PN decay in the presence of CYP102 and CPO was previously reported, but those enzymes apparently are more efficient catalysts than CYP119 for the decay of PN (9, 14, 15).
The UV-visible spectrum of CYP119-II resembled that of the CYP119 nitrosyl complex (CYP119-NO), but the gross behaviors of the spectra from the two species were significantly different. When CYP119-NO species was prepared by reaction of the resting enzyme with NO gas or with the NO donor diethylamine NONOate (diethylamine diazeniumdiolate), the UV-visible spectrum was stable for hours at room temperature, unlike the CYP119-II spectrum that decayed in seconds. This suggests that two distinct species were prepared, a conclusion that was supported by the XAS studies of the two discussed below.
The similar spectral appearance but different kinetic behavior observed for CYP119-II and CYP119-NO beg the question of how a nitrosyl complex might have been found as a product from reaction of CYP102 with PN (16). Evidence for production of a nitrosyl complex was not reported in the original stopped-flow kinetic results from reactions of CYP102 with PN (14), which resembled the results found here for reaction of CYP119 with PN, and kinetics were not followed in the more recent study (16). It seems possible that the CYP102 nitrosyl complex might be formed after PN decays because a major decomposition product from PN at neutral pH is nitrite (26, 27), and nitrite is known to react with various iron-containing enzymes (29). In studies of nitrite reactions with P450cam and CPO, the formation of nitrosyl complexes was inferred from UV-visible spectra (29–31).
As a test of the above conjecture, we briefly studied the reaction of CYP119 with sodium nitrite. When NaNO2 was added to a solution containing CYP119 in the ferric resting state, the EPR signal from the enzyme was completely lost, consistent with formation of an EPR-silent iron-nitrosyl species, and the UV-visible spectrum of the product was indistinguishable from that of authentic CYP119-NO complex (Fig. 1B). This behavior is quite similar to that reported for reactions of other ferric heme enzymes with NaNO2, which was interpreted as indicating formation of the nitrosyl complexes (29–31). The kinetics of the reaction of CYP119 with NaNO2 were complex and apparently dependent on the history of the NaNO2, suggesting that they were influenced by the presence of small amounts of impurities, but even a “fast” reaction of NaNO2 with CYP119 was 2 or 3 orders of magnitude slower than reaction of PN with CYP119. More detailed kinetic studies will be required to understand the reaction fully, but these cursory results provide a rationalization for the conflicting reports about the products formed when CYP102 was treated with PN.
Fe K-edge XAS studies were performed at Beamline 12BM of the Advanced Photon Source, Argonne National Laboratory. Samples of native CYP119, CYP119-II, and CYP119-NO complex were prepared in buffer solutions containing ≈15% ethylene glycol to prevent the formation of ice crystals that interfere with XAS signals. The enzyme concentrations were 0.3 mM for native CYP119, 0.7 mM for CYP119-II, and 1.4 mM for CYP119-NO. Samples of ≈0.2-ml volume in Kapton capillary tubes with 50-μm wall thickness were prepared at 0°C, cooled in liquid nitrogen, and mounted in a cell holder placed in line with the x-ray beam. The samples were cooled by a steady stream of cold nitrogen gas and maintained at temperatures below 140 (K) as measured by a thermocouple adjacent to the sample cell.
In acquiring the XAS spectra, the Fe K-edge position was monitored for photoreduction for each sample. No photoreduction was observed for native CYP119 and CYP119-II samples after several hours of data accumulation, whereas photoreduction was observed for the CYP119-NO samples after ≈1 h. Therefore, the CYP119-NO samples were adjusted to new positions after four scans (≈20 min each) were collected. To minimize photoreduction effects (an energy shift of the Fe K-edge position <0.3 eV) in the averaged CYP119-NO data, only the first three scans at each sample position were used. For each averaged spectrum, a Gaussian function was removed from the data to eliminate the residual signal from the elastically scattered beam before it was normalized to an edge jump of 1.0 at 7,875 eV.
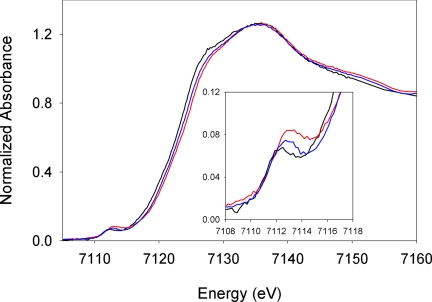
The x-ray absorption near-edge structure (XANES) spectra for native CYP119, CYP119-II, and CYP119-NO are shown in Fig. 2. A distinct shift in the edge position can easily be seen, with the edge positions of CYP119-NO and CYP119-II ≈0.4 eV and 0.8 eV, respectively, above that of the native protein. The shift observed between native CYP119 and CYP119-II is similar to that reported for native CPO and CPO-II (17). Shifts in the rising edge position are ascribed to changes in effective nuclear charge of the metal ion, and increases in the Fe K-edge energy of ≈1 eV typically signify an increase in the oxidation state by one unit (32, 33).
Fig. 2.
XANES spectra for native CYP119 (black), CYP119-II (red), and CYP119-NO (blue). (Inset) Expansion of the region containing the pre-edge features.
Table 1 lists energies and intensities of fits to the pre-edge features. Pre-edge intensities have been shown to be due to both relatively weak 1s → 3d dipole-forbidden and quadrupole-allowed transitions and strong dipole-allowed 1s → 4p transitions. In noncentrosymmetric environments, the metal 4p and 3d orbitals may mix, imparting significant pre-edge intensity. For square-pyramidal heme complexes concerned here, the greater the distortion of the metal coordination from the octahedral central-symmetry, the greater the intensity of the pre-edge feature due to the d-p mixing. All three complexes exhibit pre-edge intensities indicative of five-coordinate or highly distorted six-coordinate complexes (34): compound II (13.3 units), the NO complex (13.9 units), and the native protein (13.1 units). In these axially distorted ligand fields, the metal 4pz orbital will mix with the 3dz2 orbital, and therefore the majority of the intensity will be observed with the 3dz2 orbital. The strong-field nitrosyl and oxo ligands raise the energy of the 3dz2 orbital relative to the native complex, elevating the energy of the observed pre-edge feature along the same series as the rising edge: native CYP119 < CYP119-NO < CYP119-II (34).
Table 1.
Energies and intensities for pre-edge features in XANES spectra
| Species | 1s → 3d energy (eV) | 1s → 3d intensity, %* |
|---|---|---|
| Native CYP119 | 7112.3 ± 0.1 | 13.1 ± 2.6 |
| CYP119-II | 7112.8 ± 0.0 | 13.3 ± 1.9 |
| CYP119-NO | 7112.6 ± 0.0 | 13.9 ± 1.0 |
Errors are 1 SD.
*Percentage of absorbance normalized to 100% at 7,875 eV.
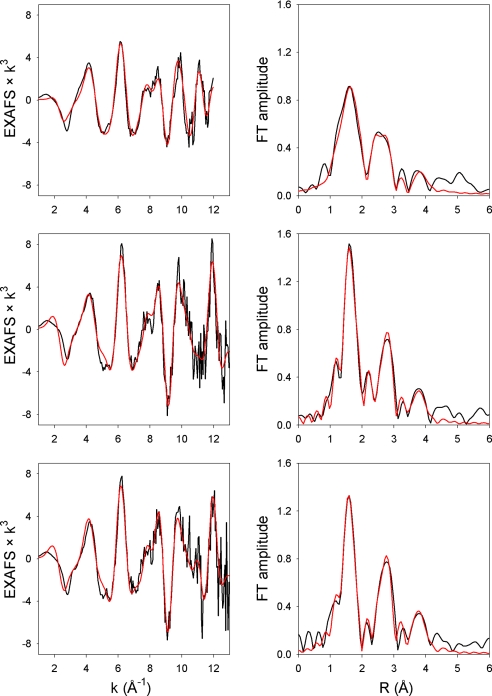
The extended x-ray absorption fine structure (EXAFS) data for native CYP119, CYP119-II, and CYP119-NO are shown in Fig. 3 along with the best fit to each dataset. The native CYP119 data are best fit (Table 2) with four nitrogen atoms of the porphyrin macrocycle at 2.00 Å and one sulfur atom from the thiolate ligand at 2.21 Å, in good agreement with crystal structure data (35). The EXAFS data for CYP119 compound II species are best fit (Table 2) with four nitrogen atoms of the porphyrin macrocycle at 2.01 Å, one sulfur atom from the thiolate ligand at 2.24 Å, and the oxo moiety oxygen at 1.82 Å. The EXAFS data for CYP119-NO complex are best fit (Table 2) with four nitrogen atoms of the porphyrin macrocycle at 1.99 Å, one sulfur atom from the thiolate ligand at 2.24 Å, and one nitrogen atom from the nitrosyl ligand at 1.82 Å. Unfortunately, the Fe-N-O multiple scattering from the nitrosyl could not be unambiguously isolated, and, thus, the Fe-N-O angle could not be determined adequately. It should be noted that in each case, three additional contributions were required to complete the fit to the more distant features in the Fourier transform: Fe-Cα single scattering, Fe-N-Cα multiple scattering, and Fe-N/C-Cβ multiple scattering. In addition, for the native CYP119 protein, a fourth additional path (Fe-Cβ single scattering) also was required. Complete parameterization for the best fits—including errors, disorder parameters, and all distances—is given in supporting information (SI) Table 3.
Fig. 3.
EXAFS results (black lines) and fits (red lines) for native CYP119 (A), CYP119-II (B), and CYP119-NO (C).
Table 2.
EXAFS bond lengths (Å) for selected porphyrin-iron-oxo and -nitrosyl species
| Species | Fe-N* | Fe-O† | Fe-S‡ | Reference |
|---|---|---|---|---|
| Native CYP119 | 2.00 | 2.21 | This work | |
| CYP119-II | 2.01 | 1.82 | 2.24 | This work |
| Native CPO | 2.02 | 2.24 | 14 | |
| CPO-II | 2.00 | 1.82 | 2.40 | 14 |
| CPO-I | 2.01 | 1.65 | 2.48 | 34 |
| (TTP)FeIV(O)(N-MeIm) | 2.03 | 1.64 | 33 | |
| (TMP+•)FeIV(O)(CH3OH) | 2.03 | 1.64 | 33 | |
| (TMP+•)FeIV(O)(Cl) | 1.99 | 1.66 | 35 | |
| (TMP+•)FeIV(O)(Br) | 2.02 | 1.65 | 35 | |
| HRP-II | 2.00 | 1.64 | 33 | |
| HRP-II | 2.00 | 1.93 | 36 | |
| HRP-II | 1.99 | 1.70 | 14 | |
| HRP-I | 2.01 | 1.65 | 14, 33, 36 | |
| Myoglobin peroxide | 1.98 | 1.69 | 37 | |
| CCP ES | 2.02 | 1.67 | 38 | |
| CYP119-NO | 1.99 | 1.82§ | 2.24 | This work |
| P450cam-NO | 2.00 | 1.76§ | 2.26 | 39 |
| P450nor-NO | 2.00 | 1.66§ | 2.26 | 39 |
CPO, chloroperoxidase; TTP, 5,10,15,20-tetra-m-tolylporphyrinato; N-MeIm, N-methylimidazole; TMP, 5,10,15,20-tetramesitylporphyrinato; HRP, horseradish peroxidase; CCP ES, cytochrome c peroxidase ES complex; P450nor, heme-thiolate nitric oxide reductase; II, compound II derivative; I, compound I derivative.
*Bond length of the four degenerate iron-nitrogen bonds from the porphyrin.
†Iron-oxygen bond length unless noted.
‡Iron-sulfur bond length for heme-thiolate proteins.
§Iron-nitrogen (of nitrosyl) bond length.
Bond lengths from the EXAFS studies of CYP119 and its derivatives are listed in Table 2, which also contains previously reported EXAFS results for porphyrin-iron-oxo species (17, 36–41) and heme-thiolate nitrosyl complexes (42). We note that the errors in EXAFS bond lengths typically are 0.01–0.02 Å. The important features of the results for CYP119-II are the long iron oxygen bond (1.81 Å) and the relatively short iron-sulfur bond (2.23 Å). Both bond lengths suggest that CYP119-II contains a protonated iron(IV) hydroxide structure as opposed to a classical ferryl group with an iron-oxygen double bond. The iron-oxygen bond length for CYP119-II is similar to that reported for CPO-II, which supported the protonated Fe-OH moiety in that species (17), but a subsequent report claimed that the sample of CPO-II contained a mixture of protonated and unprotonated species (19).
The iron-oxygen bond lengths in ferryl-oxo species correlate with bond order. For compound I species, short iron-oxo bonds (1.64–1.67 Å) were found for the compound I derivative of CPO (37), the compound I derivative of horseradish peroxidase (17, 36, 39), and the compound I model compounds, 5,10,15,20-tetramesitylporphyrin-iron(IV)-oxo radical cations (36, 38). Thus, a short, ≈1.65-Å iron-oxygen bond length is the accepted value for ferryl-oxo moieties in compound I species.
The expected bond length for a ferryl-oxo moiety in a compound II derivative is confused somewhat. Conflicting EXAFS results exist for the iron-oxygen bond length in the compound II derivative of horseradish peroxidase, as shown in Table 2 (17, 36, 39). EXAFS studies of other compound II analogues indicated short iron-oxygen bonds in a model complex (36), in myoglobin peroxide (40), and in cytochrome c peroxidase ES complex (41), each of which has a ferryl-oxo moiety without a porphyrin radical cation. Long iron-oxygen bonds (1.84–1.92 Å) were reported from x-ray crystal structure studies of the compound II derivatives of HRP (43), myoglobin (44), and catalase (45) and also the compound I derivative of cytochrome c peroxidase (46), but Green (47) noted that the x-ray crystal structure results are in conflict with EXAFS, Raman spectral, and computational results and suggested that the long iron-oxygen bonds might have resulted from reduction of the ferryl species to ferric or ferrous species in the x-ray beam.
The conclusion from previous EXAFS results is that the iron-oxygen bond length for the ferryl group in a typical compound II derivative should be somewhat less than 1.7 Å. The relatively long (1.81 Å) iron-oxygen bond in CYP119-II indicates that this compound II derivative likely contains an iron-hydroxide moiety. It is in the range of bond lengths predicted computationally for a porphyrin-iron(IV) hydroxide species (48, 49), and long iron-oxygen bonds have long been thought to be the signature of protonated ferryl-oxo species in compound II species (36, 39, 47). Accordingly, one concludes that the ferryl-oxo species in CYP119-II is basic such that the oxygen atom is protonated at neutral pH. The same conclusion was reached for CPO on the basis of the long iron-oxygen bond found for CPO-II (17). The strong basicity of the ferryl-oxo group in these compound II derivatives is a function of electron donation by the thiolate ligand, the defining feature of the P450 enzymes. Multiple effects can be ascribed to the thiolate ligand in P450s; it is thought to influence the nature of the decomposition of the transient that precedes the active oxidants in the P450 enzymes (i.e., whether the precursor cleaves by homolytic or heterolytic fragmentation of an O-O bond) (3), and it activates the oxidant toward oxygen transfer reactions by weakening the Fe-O bond (50). The thiolate ligand also strengthens the O-H bond of the Fe-OH moiety in the compound II derivative and thus reduces the acidity of these ferryl species. The strong O-H bond energy in the compound II derivative illustrates the driving force for the reactions of the more highly oxidized P450 oxidants, which can oxidize carbon-hydrogen bonds in substrates to give alcohol products.
The other interesting EXAFS result for CYP119-II is the iron-sulfur bond length, 2.23 Å. This value is similar to that found for the native enzyme, whereas the iron-sulfur bond length in the compound II derivative of CPO was longer than that in the resting enzyme and similar to that in the CPO compound I derivative (17, 37). The short iron-sulfur bond length in CYP119-II provides supporting evidence for a protonated ferryl oxygen. It reflects greater positive charge on the iron atom in the iron(IV)-hydroxide structure in comparison with the iron(IV)-oxo structure, and it is consistent with the trend in computational results (17) that predicted a 2.35-Å iron-sulfur bond for the protonated iron(IV) hydroxide structure of CPO compound II derivative and a 2.54-Å iron-sulfur bond length for the unprotonated form.
XAS studies of the CYP119-NO complex were performed largely to determine whether the species formed from the reaction of CYP119 with PN was the same as the NO complex. The EXAFS bond lengths found for CYP119-NO are similar to those for CYP119-II as expected, but the XANES spectra edge and pre-edge energies are not. The XANES spectral results agree with the UV-visible spectroscopy kinetic results that indicate that distinct species were involved. We cannot exclude the possibility that our CYP119-II samples contain some CYP119-NO, but, if the nitrosyl complex is a contaminant in CYP119-II, then it must be a minor contaminant. The XANES results for the two species are much different, and the obvious photoreduction observed during the CYP119-NO XAS data collection was not observed after hours of irradiation of the CYP119-II samples.
The interesting feature of the CYP119-NO EXAFS results is the relatively long (1.82 Å) iron-nitrogen (nitrosyl) bond. The corresponding bond length in the nitrosyl complex of the heme-thiolate nitric oxide reductase enzyme (P450nor) is 1.66 Å (42), and x-ray crystallographic studies of a model porphyrin nitrosyl derivative with a trans-thiolate ligand gave an iron-nitrogen bond length of 1.67 Å (51). The CYP119-NO result is, however, similar to that for the P450cam-NO complex, which has a 1.76-Å iron-nitrogen bond (42). A long iron-nitrogen (nitrosyl) bond length for CYP119-NO implies a significantly bent Fe-N-O bond, which is consistent with our inability to isolate multiple scattering from the nitrosyl oxygen atom (52).
In summary, XAS studies confirmed that the compound II derivative of CYP119 was produced by reaction of the resting enzyme with peroxynitrite (9) and provided a detailed description of the iron atom and its immediate surroundings in the iron(IV) derivative of this important class of enzymes. Contrary to the common view, the compound II derivative appears to be a porphyrin-iron(IV) hydroxide species as opposed to an iron-oxo species. This feature was previously found in the related heme-thiolate enzyme CPO (17) and appears to be a general consequence of the electron donation to iron by the thiolate ligand. A strong hydroxide bond in the compound II derivative of P450 enzymes demonstrates the origin of the driving force for oxidations of high-energy carbon-hydrogen bonds in substrates by the active P450 oxidants, whether they be ferryl-oxo porphyrin radical cations or perferryl-oxo species (11).
Materials and Methods
Sample Preparations.
CYP119 expression in E. coli and purification are described in refs. 22 and 53, and details of the method used here are reported in ref. 9. All samples of CYP119 used in XAS studies had R/Z values ≥1.4. Samples of CYP119-II for XAS studies were prepared by mixing at 0–5°C equal amounts of an ≈1.4 mM solution of CYP119 in 500 mM potassium phosphate buffer (pH 7.0) in ≈70:30 water-ethylene glycol with a basic solution of sodium peroxynitrate (≈120 mM) prepared from isoamyl nitrite by the method reported in ref. 24; the pH of the mixture was 7.4. A portion of the sample (≈0.2 ml) was rapidly transferred via pipette to a Kapton tube that was quickly submersed in liquid nitrogen. The remainder of the sample was transferred to a 2-mm-path-width UV cuvette, placed in a cell holder at 0°C, and analyzed by UV-visible spectroscopy to ensure that conversion to the compound II derivative was complete. The UV-visible spectrum of CYP119-II is reported in ref. 9. Samples of CYP119-NO used in XAS studies were prepared by adding commercial diethylamine NONOate as a solid to a 1.4 mM CYP119 solution at room temperature; the samples were analyzed by UV-visible spectroscopy to ensure complete conversion to the NO complex (λmax = 433 nm) before being cooled.
Kinetic Studies.
Kinetic studies of reactions of PN were performed with an SX-18MV stopped-flow mixing unit affixed to an LKS-60 kinetic spectrometer (Applied Photophysics) with single-wavelength monitoring employing a photomultiplier tube. Studies of reactions with NaNO2 were performed on an Agilent 8453 spectrophotometer with diode array detection.
Iron K-edge XAS data were collected on the bending magnet Beamline 12BM B with Si{111} crystals in the x-ray monochromator at the Advanced Photon Source, Argonne National Laboratory, and internally calibrated with an inline Fe foil, assigning the first inflection point to 7,111.2 eV. A sample temperature of <140 K was maintained throughout the experiments with a nitrogen flow delivered by an Oxford Cryosystems Cryostream 700 Plus system. Multiple samples were prepared for each species studied. Data (91 scans for native CYP119, 20 scans for CYP119-NO, and 27 scans for CYP119-II) were collected with a Canberra 13-element solid-state germanium detector array and then averaged. Successive scans of the Fe K-edge position were monitored for photoreduction for each sample. No photoreduction was observed for the native CYP119 and CYP119-II samples, whereas photoreduction was observed for the CYP119-NO sample. Therefore, only four scans were collected for each position of a sample of CYP119-NO, and only the first three scans from each position were used in the averages. For each averaged dataset, a Gaussian pre-edge function was removed from the data before it was normalized to an edge jump of 1.0 at 7,875 eV.
The intensities and energies of the Fe K-edge pre-edge features were quantified with the fitting program EDG_FIT, part of the program EXAFSPAK (54). All spectra were fit over six energy ranges (7,108, 7,119–7,116, 7,117, and 7,118 eV) by using pseudo-Voight lineshapes with an equal mix of Lorentzian and Gaussian contributions. Background functions were initially chosen empirically and then were allowed to vary in each fit along with the intensities, half-widths, and energies of the pre-edge features. Pre-edge areas were calculated as the full-width at half-maximum multiplied by amplitude. To quantify errors, standard deviations are reported along with the average pre-edge fit areas.
EXAFS data were extracted from the averaged datasets by fitting a three-segment spline to the normalized data between 7,130 and 7,875 eV. Although data were collected to k = 14 Å−1, EXAFS data were fit to k = 12 Å−1 for native CYP119, and to k = 13 Å−1 for CYP119-NO and CYP119-II because of the relative noise level of the data. EXAFS data were fit with EXAFSPAK (54), using phases and amplitudes computed by using the program FeFF 7.0 (55). Input parameters for the FeFF calculations were generated from several model structures with Fe-Npor distances ranging from 1.9 to 2.2 Å, Fe-S distances ranging from 2.10 to 2.40 Å, Fe-oxo distances ranging from 1.6 to 1.9 Å (for CYP119-II), and Fe-NNO distances ranging from 1.7 to 1.9 Å (for CYP119-NO), each in 0.1-Å increments. In processing the experimental data, monochromator glitches were removed by fitting a cubic spline to the EXAFS region and replacing a single point. In no case did glitch removal affect the final fitting results.
Supplementary Material
Acknowledgments.
We thank Nadia Leyarovska and Charles Kurtz of the Advanced Photon Source for technical assistance. This work was supported by National Institutes of Health Grant GM-48722 (to M.N.) and the Chemical Sciences Program, Basic Energy Sciences, U.S. Department of Energy, under Contract DE-AC02-06CH11357 (E.C.W. and L.X.C.). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708299105/DC1.
References
- 1.Ortiz de Montellano PR, editor. Cytochrome P450 Structure, Mechanism, and Biochemistry. 3rd Ed. New York: Kluwer; 2005. [Google Scholar]
- 2.Sono M, Roach MP, Coulter ED, Dawson JH. Chem Rev. 1996;96:2841–2887. doi: 10.1021/cr9500500. [DOI] [PubMed] [Google Scholar]
- 3.Denisov IG, Makris TM, Sligar SG, Schlichting I. Chem Rev. 2005;105:2253–2277. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]
- 4.Davydov R, Macdonald IDG, Makris TM, Sligar SG, Hoffman BM. J Am Chem Soc. 1999;121:10654–10655. [Google Scholar]
- 5.Davydov R, Perera R, Jin SX, Yang TC, Bryson TA, Sono M, Dawson JH, Hoffman BM. J Am Chem Soc. 2005;127:1403–1413. doi: 10.1021/ja045351i. [DOI] [PubMed] [Google Scholar]
- 6.Egawa T, Shimada H, Ishimura Y. Biochem Biophys Res Commun. 1994;201:1464–1469. doi: 10.1006/bbrc.1994.1868. [DOI] [PubMed] [Google Scholar]
- 7.Kellner DG, Hung SC, Weiss KE, Sligar SG. J Biol Chem. 2002;277:9641–9644. doi: 10.1074/jbc.C100745200. [DOI] [PubMed] [Google Scholar]
- 8.Spolitak T, Dawson JH, Ballou DP. J Biol Chem. 2005;280:20300–20309. doi: 10.1074/jbc.M501761200. [DOI] [PubMed] [Google Scholar]
- 9.Newcomb M, Zhang R, Chandrasena REP, Halgrimson JA, Horner JH, Makris TM, Sligar SG. J Am Chem Soc. 2006;128:4580–4581. doi: 10.1021/ja060048y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newcomb M, Hollenberg PF, Coon MJ. Arch Biochem Biophys. 2003;409:72–79. doi: 10.1016/s0003-9861(02)00445-9. [DOI] [PubMed] [Google Scholar]
- 11.Newcomb M, Chandrasena REP. Biochem Biophys Res Commun. 2005;338:394–403. doi: 10.1016/j.bbrc.2005.08.208. [DOI] [PubMed] [Google Scholar]
- 12.Zhang R, Chandrasena REP, Martinez E, Horner JH, Newcomb M. Org Lett. 2005;7:1193–1195. doi: 10.1021/ol050296j. [DOI] [PubMed] [Google Scholar]
- 13.Mehl M, Daiber A, Herold S, Shoun H, Ullrich V. Nitric Oxide Biol Chem. 1999;3:142–152. doi: 10.1006/niox.1999.0217. [DOI] [PubMed] [Google Scholar]
- 14.Daiber A, Herold S, Schöneich C, Namgaladze D, Peterson JA, Ullrich V. Eur J Biochem. 2000;267:6729–6739. doi: 10.1046/j.1432-1033.2000.01768.x. [DOI] [PubMed] [Google Scholar]
- 15.Gebicka L, Didik J. J Inorg Biochem. 2007;101:159–164. doi: 10.1016/j.jinorgbio.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 16.Behan RK, Hoffart LM, Stone KL, Krebs C, Green MT. J Am Chem Soc. 2007;129:5855–5859. doi: 10.1021/ja064590y. [DOI] [PubMed] [Google Scholar]
- 17.Green MT, Dawson JH, Gray HB. Science. 2004;304:1653–1656. doi: 10.1126/science.1096897. [DOI] [PubMed] [Google Scholar]
- 18.Stone KL, Behan RK, Green MT. Proc Natl Acad Sci USA. 2006;103:12307–12310. doi: 10.1073/pnas.0603159103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stone KL, Hoffart LM, Behan RK, Krebs C, Green MT. J Am Chem Soc. 2006;128:6147–6153. doi: 10.1021/ja057876w. [DOI] [PubMed] [Google Scholar]
- 20.Behan RK, Hoffart LM, Stone KL, Krebs C, Green MT. J Am Chem Soc. 2006;128:11471–11474. doi: 10.1021/ja062428p. [DOI] [PubMed] [Google Scholar]
- 21.Wright RL, Harris K, Solow B, White RH, Kennelly PJ. FEBS Lett. 1996;384:235–239. doi: 10.1016/0014-5793(96)00322-5. [DOI] [PubMed] [Google Scholar]
- 22.McLean MA, Maves SA, Weiss KE, Krepich S, Sligar SG. Biochem Biophys Res Commun. 1998;252:166–172. doi: 10.1006/bbrc.1998.9584. [DOI] [PubMed] [Google Scholar]
- 23.Tschirret-Guth RA, Koo LS, Hoa GHB, Ortiz de Montellano PR. J Am Chem Soc. 2001;123:3412–3417. doi: 10.1021/ja003947+. [DOI] [PubMed] [Google Scholar]
- 24.Uppu RM, Pryor WA. Anal Biochem. 1996;236:242–249. [PubMed] [Google Scholar]
- 25.Radi R, Peluffo G, Alvarez MN, Naviliat M, Cayota A. Free Radical Biol Med. 2001;30:463–488. doi: 10.1016/s0891-5849(00)00373-7. [DOI] [PubMed] [Google Scholar]
- 26.Bolzan RM, Cueto R, Squadrito GL, Uppu RM, Pryor WA. Methods Enzymol. 1999;301:178–187. doi: 10.1016/s0076-6879(99)01081-2. [DOI] [PubMed] [Google Scholar]
- 27.Kirsch M, Korth HG, Wensing A, Sustmann R, de Groot H. Arch Biochem Biophys. 2003;418:133–150. doi: 10.1016/j.abb.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Goldstein S, Lind J, Merényi G. Chem Rev. 2005;105:2457–2470. doi: 10.1021/cr0307087. [DOI] [PubMed] [Google Scholar]
- 29.Young LJ, Siegel LM. Biochemistry. 1988;27:2790–2800. doi: 10.1021/bi00408a020. [DOI] [PubMed] [Google Scholar]
- 30.Sono M, Dawson JH. J Biol Chem. 1982;257:5496–5502. [PubMed] [Google Scholar]
- 31.Sono M, Dawson JH, Hall K, Hager LP. Biochemistry. 1986;25:347–356. doi: 10.1021/bi00350a011. [DOI] [PubMed] [Google Scholar]
- 32.Wong J, Lytle FW, Messmer RP, Maylotte DH. Phys Rev B. 1984;30:5596–5610. [Google Scholar]
- 33.Wasinger EC, Davis MI, Pau MYM, Orville AM, Zaleski JM, Hedman B, Lipscomb JD, Hodgson KO, Solomon EI. Inorg Chem. 2003;42:365–376. doi: 10.1021/ic025906f. [DOI] [PubMed] [Google Scholar]
- 34.Westre TE, Kennepohl P, DeWitt JG, Hedman B, Hodgson KO, Solomon EI. J Am Chem Soc. 1997;119:6297–6314. [Google Scholar]
- 35.Park SY, Yamane K, Adachi S, Shiro Y, Weiss KE, Maves SA, Sligar SG. J Inorg Biochem. 2002;91:491–501. doi: 10.1016/s0162-0134(02)00446-4. [DOI] [PubMed] [Google Scholar]
- 36.Penner-Hahn JE, Smith Eble K, McMurry TJ, Renner M, Balch AL, Groves JT, Dawson JH, Hodgson KO. J Am Chem Soc. 1986;108:7819–7825. doi: 10.1021/ja00284a054. [DOI] [PubMed] [Google Scholar]
- 37.Stone KL, Behan RK, Green MT. Proc Natl Acad Sci USA. 2005;102:16563–16565. doi: 10.1073/pnas.0507069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolter T, Meyer-Klaucke W, Muther M, Mandon D, Winkler H, Trautwein AX, Weiss R. J Inorg Biochem. 2000;78:117–122. doi: 10.1016/s0162-0134(99)00217-2. [DOI] [PubMed] [Google Scholar]
- 39.Chance B, Powers L, Ching Y, Poulos T, Schonbaum GR, Yamazaki I, Paul KG. Arch Biochem Biophys. 1984;235:596–611. doi: 10.1016/0003-9861(84)90234-0. [DOI] [PubMed] [Google Scholar]
- 40.Chance M, Powers L, Kumar C, Chance B. Biochemistry. 1986;25:1259–1265. doi: 10.1021/bi00354a010. [DOI] [PubMed] [Google Scholar]
- 41.Chance M, Powers L, Poulos T, Chance B. Biochemistry. 1986;25:1266–1270. doi: 10.1021/bi00354a011. [DOI] [PubMed] [Google Scholar]
- 42.Obayashi E, Tsukamoto K, Adachi S, Takahashi S, Nomura M, Iizuka T, Shoun H, Shiro Y. J Am Chem Soc. 1997;119:7807–7816. [Google Scholar]
- 43.Berglund GI, Carlsson GH, Smith AT, Szoke H, Henriksen A, Hajdu J. Nature. 2002;417:463–468. doi: 10.1038/417463a. [DOI] [PubMed] [Google Scholar]
- 44.Hersleth HP, Dalhus B, Gorbitz CH, Andersson KK. J Biol Inorg Chem. 2002;7:299–304. doi: 10.1007/s007750100296. [DOI] [PubMed] [Google Scholar]
- 45.Murshudov GN, Grebenko AI, Brannigan JA, Antson AA, Barynin VV, Dodson GG, Dauter Z, Wilson KS, Melik-Adamyan WR. Acta Crystallogr D. 2002;58:1972–1982. doi: 10.1107/s0907444902016566. [DOI] [PubMed] [Google Scholar]
- 46.Bonagura CA, Bhaskar B, Shimizu H, Li HY, Sundaramoorthy M, McRee DE, Goodin DB, Poulos TL. Biochemistry. 2003;42:5600–5608. doi: 10.1021/bi034058c. [DOI] [PubMed] [Google Scholar]
- 47.Behan RK, Green MT. J Inorg Biochem. 2006;100:448–459. doi: 10.1016/j.jinorgbio.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 48.Rovira C. ChemPhysChem. 2005;6:1820–1826. doi: 10.1002/cphc.200400633. [DOI] [PubMed] [Google Scholar]
- 49.Green MT. J Am Chem Soc. 2006;128:1902–1906. doi: 10.1021/ja054074s. [DOI] [PubMed] [Google Scholar]
- 50.Zhang R, Nagraj N, Lansakara DSP, Hager LP, Newcomb M. Org Lett. 2006;8:2731–2734. doi: 10.1021/ol060762k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu N, Powell DR, Cheng L, Richter-Addo GB. Chem Commun. 2006:2030–2032. doi: 10.1039/b602611g. [DOI] [PubMed] [Google Scholar]
- 52.Westre TE, Dicicco A, Filipponi A, Natoli CR, Hedman B, Solomon EI, Hodgson KO. J Am Chem Soc. 1994;116:6757–6768. [Google Scholar]
- 53.Maves SA, Sligar SG. Protein Sci. 2001;10:161–168. doi: 10.1110/ps.17601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.George GN. EXAFSPAK. Stanford Linear Accelerator Center, Stanford Univ, Stanford, CA: Stanford Synchrotron Radiation Laboratory; 2000. [Google Scholar]
- 55.Zabinsky SI, Rehr JJ, Ankudinov A, Albers RC, Eller MJ. Phys Rev B. 1995;52:2995–3009. doi: 10.1103/physrevb.52.2995. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.