Abstract
Recently, it has been reported that mitochondria possess a novel pathway for nitric oxide (NO) synthesis. This pathway is induced when cells experience hypoxia, is nitrite (NO2−)-dependent, is independent of NO synthases, and is catalyzed by cytochrome c oxidase (Cco). It has been proposed that this mitochondrially produced NO is a component of hypoxic signaling and the induction of nuclear hypoxic genes. In this study, we examine the NO2−-dependent NO production in yeast engineered to contain alternative isoforms, Va or Vb, of Cco subunit V. Previous studies have shown that these isoforms have differential effects on oxygen reduction by Cco, and that their genes (COX5a and COX5b, respectively) are inversely regulated by oxygen. Here, we find that the Vb isozyme has a higher turnover rate for NO production than the Va isozyme and that the Vb isozyme produces NO at much higher oxygen concentrations than the Va isozyme. We have also found that the hypoxic genes CYC7 and OLE1 are induced to higher levels in a strain carrying the Vb isozyme than in a strain carrying the Va isozyme. Together, these results demonstrate that the subunit V isoforms have differential effects on NO2−-dependent NO production by Cco and provide further support for a role of Cco in hypoxic signaling. These findings also suggest a positive feedback mechanism in which mitochondrially produced NO induces expression of COX5b, whose protein product then functions to enhance the ability of Cco to produce NO in hypoxic/anoxic cells.
Keywords: hypoxia, mitochondria, reactive oxygen species, nitrite, yeast
It has been known for quite some time that the mitochondrial respiratory chain is capable of generating reactive oxygen species (ROS) that account for much of the oxidative stress experienced by cells (1–3). The levels of these ROS increase when electron flow through the respiratory chain is inhibited by respiratory inhibitors (4–6) or altered by uncoupling electron transport from oxidative phosphorylation (7, 8). Several studies have shown that exposure of cells and tissues to hypoxia increases ROS levels and oxidative stress (9–11). This increase in oxidative stress during exposure to hypoxia depends on a functional mitochondrial respiratory chain (10). It is currently unclear whether this increase is the result of increased generation or decreased destruction of ROS under hypoxic conditions. In yeast cells, the increase in ROS levels and oxidative stress is transient, as determined by shifting cells from normoxia to anoxia (10). During this shift, cells experience a continuum of decreasing oxygen concentrations and a transient increase in the levels of carbonylation of both mitochondrial and cytosolic protein, an increase in 8-OH-dG levels in mtDNA, and an increase in expression of the SOD1 gene. Many of the proteins that are carbonylated during a shift from normoxia to anoxia are the same proteins that are carbonylated when cells are exposed to menadione, a redox recycling agent that produces elevated intracellular levels of superoxide (12). When considered together, these findings indicate that cellular exposure to hypoxia during a shift from normoxia to anoxia results in increased levels of ROS.
The mitochondrial respiratory chain and cytochrome c oxidase (Cco) have been implicated in the induction of some hypoxic nuclear genes (hypoxic signaling) in both yeast and mammalian cells (13–16) exposed to reduced oxygen levels, and it has been proposed that mitochondrially generated ROS are involved (17–20). However, recent studies have revealed that ROS are not sufficient for the induction of hypoxic nuclear genes (R. Dirmeier and R.O.P., unpublished work), suggesting that the respiratory chain has an additional role in hypoxic signaling (21). An important hint concerning an additional role for the mitochondrion in hypoxic signaling comes from the findings that mitochondria from yeast, rat liver, and plants are capable of NO2−-dependent NO synthesis (21–26). This pathway is induced when cells experience hypoxia, is NO2−-dependent, is independent of NO synthases, requires the mitochondrial respiratory chain, and in yeast cells is affected by YHb, a flavohemoprotein that functions as a NO oxidoreductase (21). As proposed recently (21) mitochondrially produced NO functions in a signaling pathway to the nucleus by reacting with the superoxide produced by hypoxic mitochondria (10) to form peroxynitrite (ONOO−), which may directly or indirectly modify specific proteins. These protein modifications may involve tyrosine nitration or the nitrosation of one or more proteins (e.g., HIF-1α). Support for the involvement of mitochondrially produced NO in hypoxic signaling in yeast comes from the finding that: (i) an NO donor, 2,2′-(hydroxynitrohydrazono)bis-ethanimine (DETA-NO), induces the expression of yeast CYC7, a hypoxic nuclear gene, in normoxic cells, (ii) CYC7 is induced earlier and to higher levels in yhb1− cells than in YHB1 wild-type cells, (iii) there is an increase in protein-specific tyrosine nitration levels in cells exposed to hypoxia (21), and (iv) neither NO production nor the hypoxic induction of respiration-dependent hypoxic genes occurs in mutants that lack Cco (14, 21). This model also receives support from recent mammalian cells studies that have shown that cytochrome c, which is required for mitochondrial NO2−-dependent NO production, functions to stabilize HIF-1α in murine cell lines exposed to hypoxia (27).
The NO2−-dependent mitochondrial NO synthesis in yeast and rat liver mitochondria has been shown to be catalyzed by Cco (21). This activity is observed only at very low oxygen concentrations; which is interesting, because both yeast and mammalian cells respond to hypoxia by producing Cco isozymes with altered activities (28–30). This response is brought about by the differential expression of homologous subunit isoforms whose genes are inversely regulated by O2 (30–35). One of these isoforms (either Va or Vb in yeast and IV-1 or IV-2 in mammals) is required for activity of the Cco (33, 36) but is not part of its catalytic center. Under normoxic conditions the aerobic isoform (yeast Va or mammalian IV-1) is expressed, whereas under hypoxic or anoxic conditions the hypoxic isoform (yeast Vb or mammalian IV-2) is expressed (30, 34, 35, 37). Extensive studies with the yeast subunit V isoforms have revealed that the genes (COX5b and COX5a) for these proteins are switched on or off (respectively) at very low O2 concentrations (0.5–1 μM O2) (31) and that the isoforms affect the catalytic properties of Cco (28, 29). By altering an internal step in electron transfer between heme and the binuclear reaction center the hypoxic isoform, Vb, enhances the turnover number of the enzyme 3- to 4-fold, relative to the aerobic isoform, Va. The inverse regulation of these two isoforms allows cells to assemble different types of Cco isozymes in response to different O2 concentrations.
Given the recent findings that Cco can produce NO under hypoxic conditions (21) and that the O2-regulated subunit isoforms have differential effects on the oxidase activity of Cco, it was of interest to determine whether they affect its NO production as well. It was also of interest to determine whether these isoforms differentially affect the induction of hypoxic genes that require the mitochondrial respiratory chain.
Results
Characterization of Strains lacking YHb, Va, and Vb Singly or in Combination.
To ask whether the subunit V isoforms affect the NO2−-dependent NO production by Cco we first examined yeast cells that express either Va or Vb. We also constructed strains that are deleted for YHB1 alone or in combination with COX5a or COX5b. Strains JM43 and DR11 carry functional copies of COX5a and COX5b but should express mainly COX5a when grown under normoxic conditions because COX5b expression is repressed by the Rox1p/Reo1p repressor in the presence of air (38–40). DR11 is identical to JM43 except that it carries a null mutation in YHB1 (41). Strain GD5b carries a null mutation in COX5b (33). Strain DW29 is derived from GD5b and carries a null mutation in YHB1. Strain CT4F carries a null mutation in COX5a and the reo1–4 allele of REO1/ROX1 (34). This allele allows for the derepression of COX5b, but not all hypoxic genes, in normoxic cells (40). DW27 is derived from CT4F and carries a null mutation in YHB1.
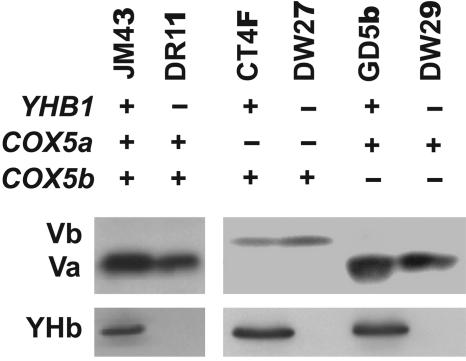
To confirm that these mutant strains contained the desired subunit V isoform, mitochondria from normoxic cells were subjected to Western immunoblot analysis (Fig. 1), using a polyclonal antibody made to a synthetic peptide constructed to correspond to the 20 aa at the carboxyl terminus common to both Va and Vb (29). Because the sequence in this region is identical in both Va and Vb, this antibody should recognize both isoforms equally well after denaturation with SDS. The Va and Vb isoforms are distinguishable on SDS gels by their mobilities: Vb migrates slightly more slowly than Va. As expected, subunit Va is present in the mitochondria of strains JM43, DR11, GD5b, and DW29 and absent in the mitochondria of strains CT4F and DW27 (Fig. 1). In contrast, subunit Vb is present in the mitochondria of strains CT4F and DW27 but absent from the other strains. Strain CT4F and its yhb1− derivative, DW27, express Vb at a reduced level relative to the level of expression of Va in strain GD5b and its yhb1− derivative, DW29.
Fig. 1.
Phenotypes of strains carrying mutations in COX5a, COX5b, or YHB1. An aliquot (30 μg) of mitochondrial protein was subjected to Western immunoblot analysis using polyclonal antibodies made to synthetic peptides corresponding to the carboxyl termini of either subunit V or YHb, as described in Materials and Methods.
To confirm the presence or absence of YHb in mitochondria from these strains we used a polyclonal antibody (42) made to a 19-aa synthetic peptide whose sequence corresponds to that predicted from the carboxyl terminus of the YHB1 gene. From Fig. 1 it is clear that YHb can be detected in mitochondria from strains JM43, CT4F, and GD5b but not in the other strains.
Effect of Subunit V Isoforms and YHb on NO2−-Dependent NO Production in Intact Yeast Cells.
The ability of the strains described above to catalyze NO2−-dependent NO synthesis was assayed under anoxic conditions (Table 1). As expected, little or no NO is produced in the two ρ0 strains, JM43ρ0 and DR10, supporting the conclusion made earlier (24) that the NO2−–dependent NO production in intact anoxic yeast cells requires a functional respiratory chain. As observed previously, the presence of YHb has a profound effect on the rate of whole-cell NO production (21). By comparing the rates of NO production in matched sets of strains [JM43 (YHB1)/DR11 (yhb1−); CT4F (YHB1)/DW27 (yhb1−); and GD5b (YHB1)/DW29 (yhb1−)] it is clear that those strains that lack YHb have rates of NO production that are between 3- and 5-fold higher than those strains that contain YHb (Table 1). It is also clear that those strains (DW27 and CT4F) with the Vb isoform have higher rates of NO2−-dependent NO production than their counterparts (DW29 and GD5b) with the Va isoform, regardless of whether YHb is present (in strains CF4F and GD5b) or absent (in strains DW27 and DW29). Given that the levels of the Vb isoform in strains CT4F and DW27 are ≈40% of those of the Va isoform in strains GD5b and DW29, the finding that strains CT4F and DW27 have higher rates of NO production than strains GD5b and DW29 is particularly striking. By comparing the rates of NO production in whole cells with their intracellular cytochrome aa3 levels, it is possible to determine the in vivo turnover rates for NO production by Cco containing either Va or Vb. These in vivo turnover rates for the Vb isozyme are 4.5 to 7 times higher than the turnover rates for the Va isozyme.
Table 1.
Effects of subunit V isozymes and YHb on NO2−-dependent NO production in yeast cells
| Strain | Subunit V isozyme | YHb | NO production, pmol NO/g wet weight per min* | In vivo turnover rate, pmol NO × 10−1/min per pmol aa3† |
|---|---|---|---|---|
| JM43 | Va | + | 78 ± 32 | 0.07 ± 0.03 |
| JM43ρ0 | Va | + | 00 ± 20 | - |
| DR11 | Va | − | 240 ± 60 | 0.21 ± 0.05 |
| DR10 | Va | − | 20 ± 10 | - |
| GD5b | Va | + | 36 ± 32 | 0.04 ± 0.03 |
| CT4F | Vb | + | 114 ± 32 | 0.30 ± 0.08 |
| DW29 | Va | − | 168 ± 40 | 0.18 ± 0.04 |
| DW27 | Vb | − | 336 ± 32 | 0.89 ± 0.08 |
*Cells (160 mg) were suspended in 2 ml of NO assay buffer and prebubbled for 5 min with N2 to create anoxic conditions. After 5 min of prebubbling NaNO2 was added to 1 mM, and NO production was measured with an NO polarographic electrode.
†Turnover rate was calculated by normalizing the rate of NO production to the cytochrome aa3 content of each strain, determined as described (31). Intracellular aa3 contents were 5.6 μM aa3/g wet weight for strains JM43 and DR11; 4.6 μM aa3/g wet weight for strains GD5b and DW29, and 1.9 μM aa3/g wet weight for strains CT4F and DW27. Strains JM43 ρ0 and DR10 lack detectable cytochromes aa3.
NO2−-Dependent NO Production in Isolated Mitochondria Carrying Va or Vb.
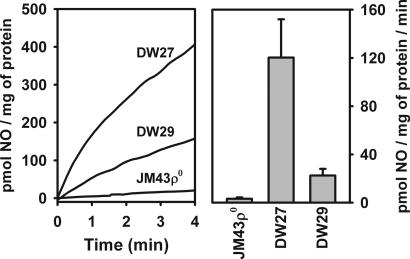
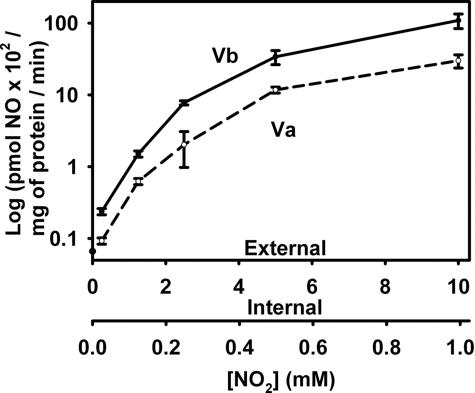
To further analyze the effects of subunit V isoforms we measured NO2−-dependent NO production in mitochondria isolated from strains JM3ρ0, DW27, and DW29. From Fig. 2Left it is clear that the rate of NO2−-dependent NO production is higher in mitochondria from a strain (DW27) carrying the Vb isozyme than in mitochondria from a strain (DW29) carrying the Va isozyme. When normalized to the level of subunit V in mitochondria (Fig. 2 Right), the rate of NO production supported by the Vb isozyme is about five times higher than the rate of the Va isozyme, which is similar to the relative rates observed above for whole cells. Mitochondria isolated from JM43ρ0 lack NO2−-dependent NO production (Fig. 2 Left). This finding is in agreement with our previous observation (21) that mitochondria from another ρ0 strain, DR10, also lacks NO2−-dependent NO production. This finding, together with the observation that the NO production observed with mitochondria from both strains DW27 and DW29 is inhibited by KCN (data not shown), supports our earlier conclusion that a functional mitochondrial respiratory chain is required for NO2−-dependent NO production in anoxic mitochondria. From Fig. 3 it is clear that increasing NO2− concentrations support higher rates of NO production by mitochondria from both DW27 and DW29 and that the rates of NO production by mitochondria from DW27, which carries the Vb isozyme, are higher than those in mitochondria from DW29, which carries the Va isozyme, over a wide range of NO2− concentrations, which fall within the physiological range (21).
Fig. 2.
NO2−-dependent NO production by mitochondria from strains DW27, DW29, and JM43ρ0. (Left) Isolated mitochondria (400 μg/ml) were suspended in NO assay medium, which was prebubbled for 5 min with N2 to create anoxic conditions. After 5 min of prebubbling NaNO2 was added to 1 mM and NO production was measured with an NO polarographic electrode. (Right) Rates of NO2−-dependent NO production by mitochondria are normalized to the subunit V content of each strain shown in Left. Mean and standard deviation values are for three independent measurements.
Fig. 3.
Effect of NO2− concentration on NO production by mitochondria from strains DW27 and DW29. NO production was measured, as described in the legend to Fig. 2, in the presence of different concentrations of added NO2− (the external concentration). The internal concentration of NO2− was measured as described in Materials and Methods. Mean and standard deviation values are for three independent measurements. The subunit V isozymes present in each mitochondrial preparation are as indicated.
Effects of O2 on NO2−-Dependent NO Production in Mitochondria with Subunit V Isozymes.
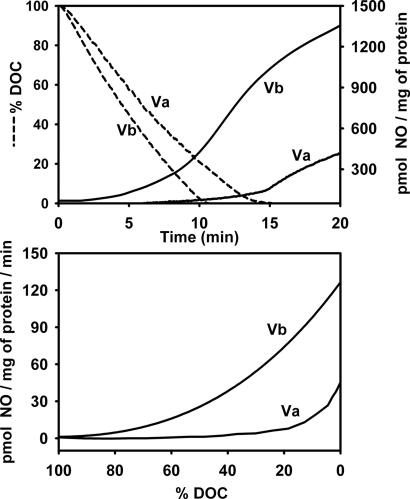
To determine whether O2 has differential effects on NO2−-dependent NO production by the Va and Vb isozymes we examined NO production over a range of O2 concentrations. These studies were done in a closed chamber in which NO production and dissolved O2 concentrations were measured concomitantly. From Fig. 4Upper it is clear that mitochondria carrying the Vb isozyme are capable of NO2−-dependent NO production at much higher O2 concentrations than mitochondria with the Va isozyme. Indeed, measurable NO production was observed in mitochondria with the Vb isozyme at a dissolved O2 concentrations as high as 80% (160 μM O2 under our assay conditions), whereas NO production in mitochondria carrying the Va isozyme is not observable until the oxygen concentrations drops <10% (20 μM O2 under our assay conditions) (Fig. 4 Lower). These findings indicate that O2 is a better inhibitor of NO2−-dependent NO production by Cco carrying Va than Vb.
Fig. 4.
Effects of oxygen concentration on NO2−-dependent NO production in mitochondria carrying different subunit V isozymes. Isolated mitochondria from strains DW27 and DW29 were assayed for NO production in assay buffer prebubbled with air in the presence of 1 mM NO2−. (Upper) NO production was measured with an NO electrode, and dissolved oxygen concentration (DOC) was measured concomitantly with an O2 electrode. (Lower) Shown are the rates of NO production as a function of DOC, as determined from the data shown in Upper. The subunit V isozymes present in each mitochondrial preparation are indicated.
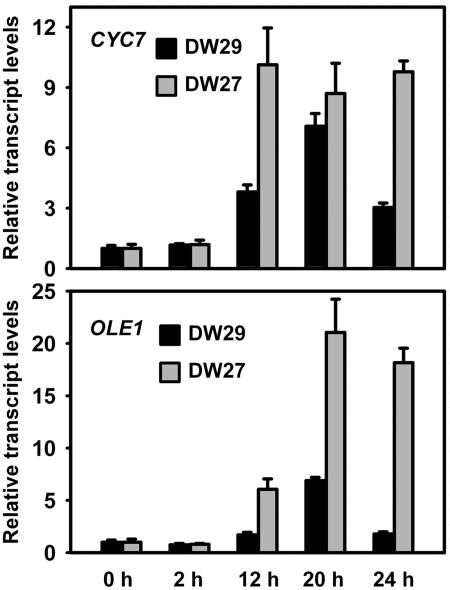
Hypoxic Induction of CYC7 and OLE1 Is Altered by Subunit V Isozymes.
Previously, it has been demonstrated that induction of the hypoxic nuclear genes CYC7 and OLE1 in yeast cells requires Cco and the respiratory chain (14). It has also been shown that DETA-NO induces the expression of CYC7 in normoxic cells and that the kinetics of induction of CYC7 in hypoxic cells is accelerated when YHB1 is deleted (21). These findings, together with the finding that Cco functions to produce NO under hypoxic and anoxic conditions, suggest that mitochondrially generated NO functions early in a hypoxic signaling pathway that is involved in hypoxic gene induction. To provide more direct support for a link between the NO production by Cco and hypoxic gene induction we asked whether the subunit V isoforms affect the induction of hypoxic genes CYC7 and OLE1. If the NO produced by Cco is involved in hypoxic gene induction one would expect that the level of hypoxic gene induction in DW27 and DW29 cells would be different, given that the Vb isozyme has a higher rate of NO2−-dependent NO production and produces NO at higher concentrations of O2 than the Va isozyme. From the data in Fig. 5 it is clear that this is the case. After a shift from normoxia to anoxia strain DW27 supports a much higher level of expression for both CYC7 and OLE1 than DW29. This difference lasts for at least 24 h after the shift.
Fig. 5.
Hypoxic induction of CYC7 and OLE1 in strains carrying different subunit V isozymes. DW27 and DW29 cells were grown for six generations in midlogarithmic phase to steady state under normoxic conditions in a fermentor. The sparge gas was then changed from air to 97.5% N2, 2.5% CO2. Cells were harvested at the times indicated, their RNA was isolated, and gene expression was analyzed by Q-PCR using primers for CYC7, OLE1, and ACT1. Transcript levels from CYC7 and OLE1 were normalized to the transcript from ACT1, whose expression is not affected by O2, and are expressed relative to their values at the beginning of the shift from normoxia to anoxia. Mean and standard deviation values are for three independent measurements.
Discussion
Cco as a NO2− Reductase.
The finding that Cco functions to produce NO• from NO2− under hypoxic conditions places it in a group with other hemoproteins that bind O2 under normoxic conditions and function in NO2−-dependent NO production under hypoxic conditions. This group includes hemoglobin and myoglobin (43–47). The rate of NO production by Cco increases with decreasing pH and increasing NO2− concentration (21), suggesting that that Cco functions in a nitrite reductase reaction (NO2− + FeII + H+ → NO + FeIII + OH−) similar to that proposed for hemoglobin and myoglobin (48–52). Unlike the Cco oxidase reaction, which requires four electrons, the Cco NO2− reductase reaction involves a one-electron transfer. Although the precise mechanism of the NO2− reduction in Cco remains to be determined, it is useful to note that a NO2−-ferric a3 complex has been observed in studies aimed at examining the interaction between NO and Cco (53–56). Moreover, it has been reported that a heme–nitrosyl complex is formed at the binuclear reaction center in bovine heart Cco when incubated in the presence of excess NO2− and reductant (57). These findings suggest that the one-electron reduction of NO2− to NO occurs at the binuclear reaction center and raise the possibility that under anoxic/hypoxic conditions a step in the catalytic cycle [e.g., NO• + compound P (Fea34+ = O2− CuB2+) → Fea3+CuB − NO2− → NO2−] (58) that is involved in the generation of NO2− from NO is merely reversed during the reduction of NO2− to NO, and that the reversal of this reaction is favored at low oxygen levels or the low pHs experienced by anoxic cells (59, 60).
Differential Effects of Subunit V Isoforms.
The finding here that the Vb isozyme supports a higher level of NO2−-dependent NO production than the Va isozyme has relevance to the evolution of Cco. Previous studies have concluded that COX5a arose from COX5b by a gene duplication event that occurred at least 130 million years ago (61). This conclusion makes it likely that oxidase activity of Cco evolved from its NO2− reductase activity and supports the conclusion that Cco arose, before atmospheric oxygen, from an enzyme whose primary function was in nitrogen metabolism, as suggested by Saraste and coworkers (62, 63). Our finding that O2 is a much better inhibitor of the Va isozyme than the Vb isozyme suggests that Va evolved in a way that serves to maximize the ability of Cco to function as an oxidase and conversely, to minimize its ability to produce NO under normoxic conditions.
Previous studies on the differential functions of the Va and Vb isozymes in yeast Cco have revealed that the rate of electron transfer from heme a to the binuclear reaction center in the Vb isozyme is 4-fold higher than it is in the Va isozyme but that the activation energies for the two isozymes are the same (28). These findings suggested that the subunit V isoforms alter the protein environment around the binuclear reaction center in such a way as to limit heme Fe2+a3's physical accessibility to electrons but without altering the barrier height of the electron transfer reaction. Fourier transform infrared analysis of carbon monoxide liganded to heme Fe2+a3 has been used to further probe the binuclear reaction centers in the subunit V isozymes (29, 64). These studies have revealed a single conformer in the Vb isozyme but two distinct conformers in the Va isozyme, supporting the conclusion that the environment around the binuclear center is different in the Va and Vb isozymes. Modeling studies have suggested that this altered rate occurs via an interaction between transmembrane helix XII of subunit I and the transmembrane helix of subunit V (32). Further insight concerning the effects of the two subunit V isoforms on the binuclear reaction center and the Cco NO2− reductase activity can be gained from analysis of their NO2−-ferric heme a3 and nitrosyl complexes, as well as the photolability of the heme a32+–NO complexes of both isozymes to different wavelengths of light (56).
Subunit V Isozymes and Hypoxic Signaling.
Until recently, the only known function of mitochondrial Cco was in the reduction of dioxygen to water coupled with proton pumping during cellular respiration. Insofar as oxygen is the electron acceptor for this process, it was surprising that gene expression studies with yeast revealed that most COX genes are expressed, albeit at low levels, under anoxic conditions (31, 65). Moreover, the promitochondria from anoxic yeast cells retain significant levels of the polypeptide subunits of Cco (66) and the ability to respire (67). These findings suggested a physiological role for Cco in the absence of O2. The recent finding that Cco functions as a NO2− reductase in the presence of reduced oxygen concentrations suggests that this physiological role is to produce NO in hypoxic or anoxic cells. This NO production is likely to be important for physiological adaptation to hypoxia, which requires the induction of several hypoxic nuclear genes (68, 69). In support of NO involvement in hypoxic signaling are the findings that: (i) the respiratory chain and Cco have been implicated in the induction of hypoxic genes in both yeast and mammalian cells (13, 14); (ii) NO production and tyrosine protein nitration increase in hypoxic yeast cells (21) in a respiration-dependent fashion; (iii) NO has a stabilizing effect on HIF-1α in mammalian cells (70), and (iv) some hypoxic yeast genes in normoxic cells are induced by exogenous NO donors (21). The finding here that the two subunit V isoforms have differential effects on the Cco NO production and on the expression of two different hypoxic genes that have been shown previously to be under the control of the respiratory chain provide further evidence for a link between Cco and hypoxic gene induction.
The expression of COX5a and COX5b is tightly regulated by oxygen (31). COX5a is switched off and COX5b is switched on when the O2 concentration drops below a threshold of 0.5 μM O2. Insofar as the full induction of COX5b under hypoxic conditions depends on a respiratory chain, which contains the Cco Va isozyme (14) when cells are shifted from normoxic to anoxic conditions, it appears that the induction of COX5b and expression of its product, Vb, constitutes a positive feedback mechanism in which hypoxia induces the expression of a gene whose product is incorporated into Cco to make it better able to produce NO. It will be interesting to determine whether a similar link can be established for the oxygen-regulated isoforms of mammalian Cco (30) and hypoxic gene induction. Although it is not yet clear which NO species are involved in hypoxic signaling likely candidates include: peroxynitrite, S-nitrosothiols, and NO (71–73). These NO species can function in protein nitration and protein nitrosation.
Conclusions
The studies described here represent a step toward addressing the mechanism underlying NO production by mitochondrial Cco and the function of the oxygen-regulated subunit V isoforms in this reaction. An in-depth understanding of both mechanisms is crucial to our understanding of hypoxic signaling and the role of NO in the regulation of respiration.
Materials and Methods
Strains, Media, and Growth Conditions.
The Saccharomyces cerevisiae strains used for this study were: JM43 (MATα his4–580 trp1–289 leu2–3, 112 ura3–52 [ρ+]) (36); JM43ρ0 (MATα his4–580 trp1–289 leu2–3, 112 ura3–52 [ρ0]) (28); DR11 (MATα his4–580 trp1–289 leu2–3, 112 ura3–52, yhb1::URA3 [ρ+]) (41); DR10 (MATα his4–580 trp1–289 leu2–3, 112 ura3–52, yhb1::URA3 [ρ0]) (41); CT4F (MATα his4–580 trp1–289 leu2–3, 112 ura3–52, cox5aΔ::URA3, COX5b, reo1–4, [ρ+]) (34); GD5b (MATα his4–580 trp1–289 leu2–3, 112 ura3–52 cox5b::LEU2 [ρ+]) (34); YDW27 (MATα his4–580 trp1–289 leu2–3, 112 ura3–52, cox5aΔ::URA3, COX5b, reo1–4, yhb1::Kanr, [ρ+]); and YDW29 (MATα his4–580 trp1–289 leu2–3, 112 ura3–52 cox5b::LEU2, yhb1:: Kanr [ρ+]). All strains were derived from and are isochromosomal with JM43. Strains YDW27 and YDW29 were constructed for this study by selective deletion of the YHB1 genes in strains CT4F and GD5b, respectively, using homologous recombination of site-specific cassettes amplified by PCR (74). Deletion cassettes containing the Escherichia coli kanr gene, which confers resistance to G418/geneticin (75, 76), were used to transform CT4F and GD5b by the lithium acetate procedure (77).
Yeast cells were grown in SSG-TEA, a semisynthetic medium (66), supplemented with Tween 80, ergosterol, silicon antifoam, and amino acids and uracil, as needed (14). Aerobic cultures and precultures were grown in a controlled environment incubator shaker (New Brunswick Scientific) at 200 rpm and 28°C and harvested in midlogarithmic growth phase. Shift experiments between normoxia and anoxia were performed in a New Brunswick BIOFLO 3000 fermentor, as described (78).
Preparation of Mitochondria.
Mitochondria were prepared from aerobic cultures as described (79). After cell breakage and centrifugation at 2,000 × g for 3 min, an aliquot of the supernatant was saved as “whole-cell lysate.” The rest of the supernatant was decanted and centrifuged for 10 min at 12,000 × g to pellet the mitochondrial fraction, saving the supernatant as the cytosolic fraction. Oxygen uptake was measured at 30°C with a Strathkelvin oxygen electrode system, as described (78), but with the following modifications: the assay solution consisted of 2 ml of buffer containing 0.65 M Mannitol, 0.01M K2HPO4 (pH 6.5), 0.1 mM EDTA, 0.01 M KCl, 5 mM α-ketoglutarate, and 0.7 mg of mitochondrial protein. The P:O and respiratory control ratios of mitochondria were determined (49) by measuring oxygen consumption in the presence or absence of ADP and only coupled mitochondria (P:O ≈2) were used.
SDS/PAGE and Immunoblot Analysis.
SDS/PAGE was performed on 16% SDS polyacrylamide gels containing 10% glycerol and 3.6 M urea (29). After electrophoresis, proteins were electroblotted to Polyscreen PVDF transfer membrane (PerkinElmer), immunoblotted with a polyclonal antibody to either YHb (42) or a polyclonal antibody to the carboxyl terminal of subunit V (29), and detected with peroxidase-linked antibodies and a Western Lightning chemiluminescence detection kit (PerkinElmer). The levels of Va and Vb on immunoblots were quantitated by using ImageQuant TL software (GE Healthcare Biosciences).
Measurement of NO Production.
NO production was measured with a 2-mm Clark-type NO electrode and an APOLLO 4000 NO-meter (WPI) (21). Except where noted, all solutions were NO2− free. Measurements were performed at 28°C with a final reaction volume of 2 ml in a thermostated chamber with a close-fitting lid and fine holes for the electrode and a Hamilton syringe. Assays for NO production by the mitochondrial respiratory chain were performed in NO assay medium [6 mM succinate, 650 mM Mannitol, 10 mM K2HPO4 (pH 6.5), 0.1 mM EDTA, and 10 mM KCl]. Except where noted, the NO assay buffer was prebubbled with N2 to achieve anoxic conditions before addition of NaNO2 to a final concentration of 1 mM and the measurement of NO production.
Measurement of Mitochondrial Internal NO2− Concentration.
Evaluation of NO2− uptake by mitochondria obtained from yeast strains DW27 and DW29 was performed as described (21).
Quantitative-PCR (Q-PCR).
Total RNA was isolated from yeast cells as described (80). Q-PCR amplification reactions were performed with an iScript cDNA Synthesis Kit (BioRad) following the manufacturer's instructions. Primers were designed against sequences of the indicated mRNAs by using Primer Express 2.0 (Applied Biosystems). An aliquot of 25-μl PCRs containing 1× SYBR Green Mix (Applied Biosystems), 1 ng of cDNA, and 500 nM primers were set up in 96-well plates. Standard curves from 1–10 ng of cDNA were run alongside samples for each individual primer, and plates were read in an Applied Biosystems 7900HT Q-PCR instrument (absolute quantification method). Expression quantities were normalized to the ACT1 transcript.
Miscellaneous Methods.
Protein concentration was determined by either the Lowry assay (80) or the BCA assay (Pierce Biotechnology) with BSA as a standard.
Acknowledgments.
We thank Joaquin M. Espinosa for assistance in the quantitation of genes. This work was supported by National Institutes of Health Grant GM 30228 (to R.O.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Boveris A, Chance B. Biochem J. 1973;134:707–716. doi: 10.1042/bj1340707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cadenas E. Annu Rev Biochem. 1989;58:79–110. doi: 10.1146/annurev.bi.58.070189.000455. [DOI] [PubMed] [Google Scholar]
- 3.Turrens JF. J Physiol (London) 2003;552:335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bandy B, Davison AJ. Free Radical Biol Med. 1990;8:523–539. doi: 10.1016/0891-5849(90)90152-9. [DOI] [PubMed] [Google Scholar]
- 5.St-Pierre J, Buckingham JA, Roebuck SJ, Brand MD. J Biol Chem. 2002;277:44784–44790. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- 6.Turrens JF, Alexandre A, Lehninger AL. Arch Biochem Biophys. 1985;237:408–414. doi: 10.1016/0003-9861(85)90293-0. [DOI] [PubMed] [Google Scholar]
- 7.Brand MD, Esteves TC. Cell Metab. 2005;2:85–93. doi: 10.1016/j.cmet.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Starkov AA. Chem Biol Interact. 2006;161:57–68. doi: 10.1016/j.cbi.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 9.Chandel NS, Budinger GR. Free Radical Biol Med. 2007;42:165–174. doi: 10.1016/j.freeradbiomed.2006.10.048. [DOI] [PubMed] [Google Scholar]
- 10.Dirmeier R, O'Brien KM, Engle M, Dodd A, Spears E, Poyton RO. J Biol Chem. 2002;277:34773–34784. doi: 10.1074/jbc.M203902200. [DOI] [PubMed] [Google Scholar]
- 11.Grishko V, Solomon M, Breit JF, Killilea DW, LeDoux SP, Wilson GL, Gillespie MN. FASEB J. 2001;15:1267–1269. doi: 10.1096/fj.00-0755fje. [DOI] [PubMed] [Google Scholar]
- 12.Cabiscol E, Piulats E, Echave P, Herrero E, Ros J. J Biol Chem. 2000;275:27393–27398. doi: 10.1074/jbc.M003140200. [DOI] [PubMed] [Google Scholar]
- 13.Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, Schumacker PT. Proc Natl Acad Sci USA. 1998;95:11715–11720. doi: 10.1073/pnas.95.20.11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwast KE, Burke PV, Staahl BT, Poyton RO. Proc Natl Acad Sci USA. 1999;96:5446–5451. doi: 10.1073/pnas.96.10.5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poyton RO. Respir Physiol. 1999;115:119–133. doi: 10.1016/s0034-5687(99)00028-6. [DOI] [PubMed] [Google Scholar]
- 16.Poyton RO, Dirmeier R, O'Brien K, Spears E. In: Oxygen Sensing: Responses and Adaptation to Hypoxia. Lahiri S, Semenza GL, Prabhakar NR, editors. New York: Dekker; 2003. pp. 23–45. [Google Scholar]
- 17.Chandel NS, McClintock DS, Feliciano CE, Wood TM, Melendez JA, Rodriguez AM, Schumacker PT. J Biol Chem. 2000;275:25130–25138. doi: 10.1074/jbc.M001914200. [DOI] [PubMed] [Google Scholar]
- 18.Brunelle JK, Bell EL, Quesada NM, Vercauteren K, Tiranti V, Zeviani M, Scarpulla RC, Chandel NS. Cell Metab. 2005;1:409–414. doi: 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Guzy RD, Hoyos B, Robin E, Chen H, Liu L, Mansfield KD, Simon MC, Hammerling U, Schumacker PT. Cell Metab. 2005;1:401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 20.Kaelin WG., Jr Cell Metab. 2005;1:357–358. doi: 10.1016/j.cmet.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 21.Castello PR, David PS, McClure T, Crook Z, Poyton RO. Cell Metab. 2006;3:277–287. doi: 10.1016/j.cmet.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 22.Gupta KJ, Stoimenova M, Kaiser WM. J Exp Bot. 2005;56:2601–2609. doi: 10.1093/jxb/eri252. [DOI] [PubMed] [Google Scholar]
- 23.Nohl H, Staniek K, Sobhian B, Bahrami S, Redl H, Kozlov AV. Acta Biochim Pol. 2000;47:913–921. [PubMed] [Google Scholar]
- 24.Nohl H, Staniek K, Kozlov AV. Redox Rep. 2005;10:281–286. doi: 10.1179/135100005X83707. [DOI] [PubMed] [Google Scholar]
- 25.Planchet E, Jagadis GK, Sonoda M, Kaiser WM. Plant J. 2005;41:732–743. doi: 10.1111/j.1365-313X.2005.02335.x. [DOI] [PubMed] [Google Scholar]
- 26.Tischner R, Planchet E, Kaiser WM. FEBS Lett. 2004;576:151–155. doi: 10.1016/j.febslet.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 27.Mansfield KD, Guzy RD, Pan Y, Young RM, Cash TP, Schumacker PT, Simon MC. Cell Metab. 2005;1:393–399. doi: 10.1016/j.cmet.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waterland RA, Basu A, Chance B, Poyton RO. J Biol Chem. 1991;266:4180–4186. [PubMed] [Google Scholar]
- 29.Allen LA, Zhao XJ, Caughey W, Poyton RO. J Biol Chem. 1995;270:110–118. doi: 10.1074/jbc.270.1.110. [DOI] [PubMed] [Google Scholar]
- 30.Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV, Semenza GL. Cell. 2007;129:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 31.Burke PV, Raitt DC, Allen LA, Kellogg EA, Poyton RO. J Biol Chem. 1997;272:14705–14712. doi: 10.1074/jbc.272.23.14705. [DOI] [PubMed] [Google Scholar]
- 32.Burke PV, Poyton RO. J Exp Biol. 1998;201:1163–1175. doi: 10.1242/jeb.201.8.1163. [DOI] [PubMed] [Google Scholar]
- 33.Trueblood CE, Poyton RO. Mol Cell Biol. 1987;7:3520–3526. doi: 10.1128/mcb.7.10.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trueblood CE, Poyton RO. Genetics. 1988;120:671–680. doi: 10.1093/genetics/120.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trueblood CE, Wright RM, Poyton RO. Mol Cell Biol. 1988;8:4537–4540. doi: 10.1128/mcb.8.10.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cumsky MG, Ko C, Trueblood CE, Poyton RO. Proc Natl Acad Sci USA. 1985;82:2235–2239. doi: 10.1073/pnas.82.8.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poyton RO, Burke PV. Biochim Biophys Acta. 1992;1101:252–256. [PubMed] [Google Scholar]
- 38.Zitomer RS, Limbach MP, Rodriguez-Torres AM, Balasubramanian B, Deckert J, Snow PM. Methods. 1997;11:279–288. doi: 10.1006/meth.1996.0422. [DOI] [PubMed] [Google Scholar]
- 39.Zitomer RS, Carrico P, Deckert J. Kidney Int. 1997;51:507–513. doi: 10.1038/ki.1997.71. [DOI] [PubMed] [Google Scholar]
- 40.Kwast KE, Burke PV, Brown K, Poyton RO. Curr Genet. 1997;32:377–383. doi: 10.1007/s002940050291. [DOI] [PubMed] [Google Scholar]
- 41.Zhao XJ, Raitt D, Burke V, Clewell AS, Kwast KE, Poyton RO. J Biol Chem. 1996;271:25131–25138. doi: 10.1074/jbc.271.41.25131. [DOI] [PubMed] [Google Scholar]
- 42.Cassanova N, O'Brien KM, Stahl BT, McClure T, Poyton RO. J Biol Chem. 2005;280:7645–7653. doi: 10.1074/jbc.M411478200. [DOI] [PubMed] [Google Scholar]
- 43.Copeland DM, Soares AS, West AH, Richter-Addo GB. J Inorg Biochem. 2006;100:1413–1425. doi: 10.1016/j.jinorgbio.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 44.Cosby K, Partovi KS, Crawford JH, Patel RP, Reiter CD, Martyr S, Yang BK, Waclawiw MA, Zalos G, Xu X, et al. Nat Med. 2003;9:1498–1505. doi: 10.1038/nm954. [DOI] [PubMed] [Google Scholar]
- 45.Nagababu E, Ramasamy S, Abernethy DR, Rifkind JM. J Biol Chem. 2003;278:46349–46356. doi: 10.1074/jbc.M307572200. [DOI] [PubMed] [Google Scholar]
- 46.Reutov VP, Sorokina EG. Biochemistry (Mosc) 1998;63:874–884. [PubMed] [Google Scholar]
- 47.Shiva S, Huang Z, Grubina R, Sun J, Ringwood LA, Macarthur PH, Xu X, Murphy E, Darley-Usmar VM, Gladwin MT. Circ Res. 2007;100:654–661. doi: 10.1161/01.RES.0000260171.52224.6b. [DOI] [PubMed] [Google Scholar]
- 48.Sulc F, Immoos CE, Pervitsky D, Farmer PJ. J Am Chem Soc. 2004;126:1096–1101. doi: 10.1021/ja0376184. [DOI] [PubMed] [Google Scholar]
- 49.Doyle MP, Pickering RA, DeWeert TM, Hoekstra JW, Pater D. J Biol Chem. 1981;256:12393–12398. [PubMed] [Google Scholar]
- 50.Bryan NS. Free Radical Biol Med. 2006;41:691–701. doi: 10.1016/j.freeradbiomed.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 51.Grubina R, Huang Z, Shiva S, Joshi MS, Azarov I, Basu S, Ringwood LA, Jiang A, Hogg N, Kim-Shapiro DB, et al. J Biol Chem. 2007;282:12916–12927. doi: 10.1074/jbc.M700546200. [DOI] [PubMed] [Google Scholar]
- 52.Grubina R, Basu S, Tiso M, Kim-Shapiro DB, Gladwin MT. J Biol Chem. 2008;283:3628–3638. doi: 10.1074/jbc.M705222200. [DOI] [PubMed] [Google Scholar]
- 53.Cooper CE, Torres J, Sharpe MA, Wilson MT. FEBS Lett. 1997;414:281–284. doi: 10.1016/s0014-5793(97)01009-0. [DOI] [PubMed] [Google Scholar]
- 54.Torres J, Sharpe MA, Rosquist A, Cooper CE, Wilson MT. FEBS Lett. 2000;475:263–266. doi: 10.1016/s0014-5793(00)01682-3. [DOI] [PubMed] [Google Scholar]
- 55.Giuffre A, Barone MC, Mastronicola D, D'Itri E, Sarti P, Brunori M. Biochemistry. 2000;39:15446–15453. doi: 10.1021/bi000447k. [DOI] [PubMed] [Google Scholar]
- 56.Sarti P, Giuffre A, Forte E, Mastronicola D, Barone MC, Brunori M. Biochem Biophys Res Commun. 2000;274:183–187. doi: 10.1006/bbrc.2000.3117. [DOI] [PubMed] [Google Scholar]
- 57.Brudvig GW, Stevens TH, Chan SI. Biochemistry. 1980;19:5275–5285. doi: 10.1021/bi00564a020. [DOI] [PubMed] [Google Scholar]
- 58.Brunori M, Forte E, Arese M, Mastronicola D, Giuffre A, Sarti P. Biochim Biophys Acta. 2006;1757:1144–1154. doi: 10.1016/j.bbabio.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 59.Campbellburk SL, Denhollander JA, Alger JR, Shulman RG. Biochemistry. 1987;26:7493–7500. doi: 10.1021/bi00397a044. [DOI] [PubMed] [Google Scholar]
- 60.Gonzalez B, de Graaf A, Renaud M, Sahm H. Yeast. 2000;16:483–497. doi: 10.1002/(SICI)1097-0061(200004)16:6<483::AID-YEA542>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 61.Cumsky MG, Trueblood CE, Ko C, Poyton RO. Mol Cell Biol. 1987;7:3511–3519. doi: 10.1128/mcb.7.10.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Castresana J, Lubben M, Saraste M, Higgins DG. EMBO J. 1994;13:2516–2525. doi: 10.1002/j.1460-2075.1994.tb06541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saraste M, Castresana J. FEBS Lett. 1994;341:1–4. doi: 10.1016/0014-5793(94)80228-9. [DOI] [PubMed] [Google Scholar]
- 64.Zhao XJ, Caughey WS, Poyton RO. Methods Enzymol. 1995;260:399–406. doi: 10.1016/0076-6879(95)60153-8. [DOI] [PubMed] [Google Scholar]
- 65.Kwast KE, Burke PV, Poyton RO. J Exp Biol. 1998;201:1177–1195. doi: 10.1242/jeb.201.8.1177. [DOI] [PubMed] [Google Scholar]
- 66.Dagsgaard C, Taylor LE, O'Brien KM, Poyton RO. J Biol Chem. 2001;276:7593–7601. doi: 10.1074/jbc.M009180200. [DOI] [PubMed] [Google Scholar]
- 67.David PS, Poyton RO. Biochim Biophys Acta. 2005;1709:169–180. doi: 10.1016/j.bbabio.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 68.Bunn HF, Poyton RO. Physiol Rev. 1996;76:839–885. doi: 10.1152/physrev.1996.76.3.839. [DOI] [PubMed] [Google Scholar]
- 69.Wenger RH. J Exp Biol. 2000;203:1253–1263. doi: 10.1242/jeb.203.8.1253. [DOI] [PubMed] [Google Scholar]
- 70.Li F, Sonveaux P, Rabbani ZN, Liu S, Yan B, Huang Q, Vujaskovic Z, Dewhirst MW, Li CY. Mol Cell. 2007;26:63–74. doi: 10.1016/j.molcel.2007.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.El-Remessy AB, Al-Shabrawey M, Platt DH, Bartoli M, Behzadian MA, Ghaly N, Tsai N, Motamed K, Caldwell RB. FASEB J. 2007;21:2528–2539. doi: 10.1096/fj.06-7854com. [DOI] [PubMed] [Google Scholar]
- 72.Gaston B, Singel D, Doctor A, Stamler JS. Am J Respir Crit Care Med. 2006;173:1186–1193. doi: 10.1164/rccm.200510-1584PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Palmer LA, Doctor A, Chhabra P, Sheram ML, Laubach VE, Karlinsey MZ, Forbes MS, Macdonald T, Gaston B. J Clin Invest. 2007;117:2592–2601. doi: 10.1172/JCI29444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 75.Jimenez A, Davies J. Nature. 1980;287:869–871. doi: 10.1038/287869a0. [DOI] [PubMed] [Google Scholar]
- 76.Hadfield C, Jordan BE, Mount RC, Pretorius GH, Burak E. Curr Genet. 1990;18:303–313. doi: 10.1007/BF00318211. [DOI] [PubMed] [Google Scholar]
- 77.Ito H, Fukuda Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Poyton RO, Dirmeier R, O'Brien K, David P, Dodd A. Methods Enzymol. 2004;381:644–662. doi: 10.1016/S0076-6879(04)81042-5. [DOI] [PubMed] [Google Scholar]
- 79.McKee EE, Poyton RO. J Biol Chem. 1984;259:9320–9331. [PubMed] [Google Scholar]
- 80.Lowry OH, Rosebrough NJ, Farral AL, Randall RJ. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]