Abstract
Glycosylation is one of the most common posttranslational modifications to occur in protein biosynthesis, yet its effect on the thermodynamics and kinetics of proteins is poorly understood. A minimalist model based on the native protein topology, in which each amino acid and sugar ring was represented by a single bead, was used to study the effect of glycosylation on protein folding. We studied in silico the folding of 63 engineered SH3 domain variants that had been glycosylated with different numbers of conjugated polysaccharide chains at different sites on the protein's surface. Thermal stabilization of the protein by the polysaccharide chains was observed in proportion to the number of attached chains. Consistent with recent experimental data, the degree of thermal stabilization depended on the position of the glycosylation sites, but only very weakly on the size of the glycans. A thermodynamic analysis showed that the origin of the enhanced protein stabilization by glycosylation is destabilization of the unfolded state rather than stabilization of the folded state. The higher free energy of the unfolded state is enthalpic in origin because the bulky polysaccharide chains force the unfolded ensemble to adopt more extended conformations by prohibiting formation of a residual structure. The thermodynamic stabilization induced by glycosylation is coupled with kinetic stabilization. The effects introduced by the glycans on the biophysical properties of proteins are likely to be relevant to other protein polymeric conjugate systems that regularly occur in the cell as posttranslational modifications or for biotechnological purposes.
Keywords: energy landscape, posttranslational, modification, unfolded state
Posttranslational modifications are ubiquitous in the cell and regulate the function of proteins often by modulating their biophysical characteristics. These modifications (e.g., glycosylation, phosphorylation, and methylation) can adjust the thermodynamic, kinetic, and structural features of proteins. Posttranslational modifications, therefore, enrich the repertoire of protein characteristics beyond that dictated by their sequence and can be viewed as an economical way to introduce larger diversity to the proteins encoded in the genome.
The biophysical properties of proteins are a consequence of the features of the underlying energy landscapes that govern their folding. Joint experimental and theoretical efforts (1, 2) have concluded that protein fold by navigating through an energy landscape that is globally funneled toward a structurally defined native state (3). The funneled nature of energy landscapes is a product of an evolutionary selection of sequences that are minimally frustrated in that they have fewer interactions that are in conflict with the native state and thus exhibit relatively high conformational specificity (i.e., a smoother landscape). The funneled energy landscape for protein folding explains why protein topology so strongly determines folding and binding kinetics. Using the funneled energy landscape concept, many experimental data on protein folding thermodynamics and kinetics have been reproduced, including the molecular structure of intermediates and the ensembles of the transition state, and protein structures have been predicted and designed (4–6).
However, attaining full comprehension of isolated monomeric proteins does not endow complete understanding of the biological functioning of proteins in the cell. Understanding the effects of posttranslational modifications to the protein energy landscape is valuable in understanding protein function and how protein thermodynamics and kinetics can be modulated by the formation of a conjugate or through an external stimulus. In this article, we explore the effects of glycosylation on the biophysical properties of proteins with the main goal of understanding folding mechanisms, thermodynamics, and kinetics in the context of the cell.
Glycosylation [i.e., the attachment of polysaccharide chains (also termed “glycans”) to proteins] is regarded as one of the most common and important posttranslational modifications to occur during or after protein synthesis (7, 8). Glycosylation is a complex process that involves 13 different kinds of monosaccharides attached to eight types of amino acid residues (9) and is assisted by many different enzymes. Glycosylation is an effective way of generating diversity in proteins and modulating their properties due to the inherent structural variations of glycans. Among their repertoire of functional roles, glycans serve as recognition markers, mediate interaction with pathogens, modulate immune responses, and regulate protein turnover (8, 10). Difficulties in characterizing the glycans and synthesizing glycoproteins made this field a challenging one. New techniques of glycopeptide and carbohydrate synthesis have enabled significant progress in glycoprotein folding studies, but still the interplay between carbohydrates and protein folding kinetics and stability is unclear.
Glycosylation starts at the endoplasmic reticulum during protein synthesis in the ribosome. The glycan is added to the unfolded protein while it is in the translocon complex (11), suggesting that it may assist in obtaining the correct fold. The role of glycans in gaining the correct fold is, however, ambiguous. There is evidence for protein misfolding and aggregation in the absence of glycans, although in other cases elimination of all or some glycans has no effect on folding (see refs. 12 and 13 and references therein), which implies that some glycosylation sites are more crucial for folding than others and that the effect of glycans on folding is likely to be local. Although glycans can assist protein folding, their removal from folded proteins often does not affect the protein fold and function.
The glycans, which are bulky hydrophilic polymers, often contribute to the high solubility of the protein and increase its stability against proteolysis. Moreover, the covalent binding of glycans to the protein surface may inherently enhance the thermal and kinetic stability of proteins. They can, in principle, stabilize proteins by affecting their underlying energy landscapes. The higher stability is expressed by a higher melting temperature for the glycosylated protein and/or by the free-energy difference between the unfolded and folded state ensembles being greater for the glycosylated protein than for its nonglycosylated wild-type counterpart (i.e., ΔGGlyco>ΔGWT (see Fig. 1). Several studies of the folding and stability of glycoproteins in their glycosylated and nonglycosylated forms show that the sugars have diverse effects on the folding kinetics and its pathway (14), yet most studies indicate an enhanced thermodynamic stability in the glycosylated form. Some experimental studies suggest that stabilization by glycosylation is due to the formation of new interactions between the sugars and the amino acids in the native state (15), namely, the stabilization effect in these cases is enthalpic in origin. Several previous experimental studies (16–19) that attempted to characterize the “chaperone-like” activity of glycans suggest that glycoprotein thermal stabilization is largely entropic in origin, rather than enthalpic (11). Accordingly, there is controversy as to whether glycans affect the entropy or enthalpy and whether these effects are mainly on the folded or unfolded state.
Fig. 1.
Schematic free-energy profile of a glycoprotein. Glycosylation may result in higher stability compared with the nonglycosylated wild-type protein (i.e., ΔGGlyco>ΔGWT). The higher stability can be the outcome of a less stable unfolded state (i.e., ΔGUGlyco−ΔGUWT>0) or a more stable folded state (i.e., ΔGFGlyco−ΔGFWT<0). Glycosylation also may affect the height of the folding barrier.
Several possible changes in the energy landscape of the glycosylated proteins can explain their higher thermostability. For example, greater thermostability can be obtained by decreasing the enthalpy or increasing the entropy of the folded state. Alternatively, the overall stability of the protein can be improved by destabilizing the unfolded state via decreasing its entropy or increasing its enthalpy (e.g., by breaking some residual structural elements). Any one of these actions, or a combination of them, can explain the generally higher thermal and chemical stability of glycosylated proteins.
To explore the origin of the thermal stabilization of proteins that is gained upon glycosylation, we studied the folding mechanisms of SH3 domain carbohydrate conjugate variants obtained by combinatorial glycosylation at six selected glycosylation sites. The SH3 domain is a small protein (56 amino acids) (Fig. 2) whose folding was well studied in the past, both experimentally and theoretically. Although the SH3 domain is not a glycoprotein (i.e., it is not a realistic study case), studying a protein whose folding is well characterized can give an insight into the general effects of the glycan on folding. Two types of polysaccharides were used: one that includes 5 sugar rings and a single branch and another that includes 11 sugar rings and two branches (see Fig. 2). We added polysaccharide chains at up to six artificial glycosylation sites on the surface of the SH3 protein, and we studied the effects of the number of glycosylations and their location on the stability of the protein and its folding mechanism.
Fig. 2.
Ribbon diagram of the SH3 domain with six “designed” polysaccharide chains. The polypeptide chain is colored blue, and the carbohydrate rings are represented as gray balls. Each glycan contains 11 sugar rings. In the coarse-grained model, each sugar ring is represented by a single bead with a radius of 6 Å.
In this article, the folding mechanism of the glycoproteins was studied by using computational tools. Several computational studies of glycoproteins were previously performed with full-atom molecular dynamic simulations (20–23). In these works, either only short glycopeptides (≈10 residues) were studied (22) or the dynamics of the glycoprotein was often limited to the folded state (23). An alternative approach is to study the folding of glycoproteins by using coarse-grained models that use reduced representations of the protein and oligosaccharides. Here, we applied a native topology-based (Go) model. In this model, each amino acid and sugar ring was represented by a single bead. This approach allowed us to study the folding of a protein having a complex topology and to investigate the effect of the glycans on the entropy and enthalpy of the various states involved in the folding reaction.
Consistent with the available experimental data, we found that the glycans increase the stability of the protein. A detailed thermodynamic analysis shows that this stabilization is achieved primarily by increasing the enthalpy of the unfolded state.
Results and Discussion
We designed in silico artificial glycosylated variants of the SH3 domain protein. Analysis of structures of glycoproteins in the Protein Data Bank has suggested that glycosylation sites usually appear in sites of alternating secondary structure motifs (24). Accordingly, for the SH3 protein, we chose six glycosylation sites that are located on loops and coils and that do not disturb the protein structure when glycosylated. We designed all possible glycosylated variants obtainable from the 5- and 11-sugar ring polysaccharides that we used (Fig. 2). This design approach resulted in 63 glycosylated variants for each type of sugar: 6 variants with a single glycan (at each of the six glycosylation sites), 15 with two glycans, 20 with three glycans, 15 with four glycans, 6 with five glycans, and a single fully glycosylated variant with all of the six positions being glycosylated. The folding of each of the glycosylated forms of the SH3 domain was characterized by using the native topology-based model. The simulations, which covered a broad range of temperatures, were performed at equilibrium so that each trajectory included several folding/unfolding transitions. From these simulations, the thermodynamic and kinetic features of each variant were obtained, providing a good starting point to look for correlations between the degree and location of glycosylation and the biophysical properties of proteins.
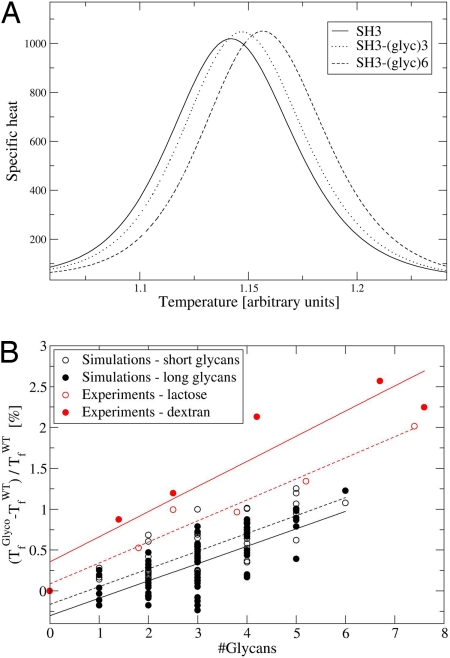
Fig. 3A shows the specific heats of SH3 (solid line) and two of its glycosylated variants, one bearing three glycans and the other six glycans. The peak of the curve represents the folding temperature (Tf) of the protein at which the free energies of the folded and unfolded states are very similar (i.e., ΔGGlyco = ΔGWT≈0). It can be observed that Tf increases as the degree of glycosylation increases. Fig. 3B summarizes the Tf values calculated for all 63 glycosylated SH3 variants [those conjugated with the 11-sugar ring glycan (filled circles) and those conjugated with the 5-sugar ring glycan (open circles)]. Our simulations indicate that protein stability increases as the degree of glycosylation increases. This observation is in accordance with the available experimental data. In general, experiments demonstrated that glycans increase the thermal stability of the protein (18, 25, 26). In particular, Sola et al. (27, 28) demonstrated that the chemical glycosylation of α-cymotrypsin (α-CT) using a lactose comprising two sugar rings and a dextran of 10 kDa increased the melting temperature of the protein in a manner that depends on the amount of glycan involved. They performed a systematic chemical glycosylation of α-CT on all the surface-exposed lysine residues with increasing concentrations of glycan. Their results also are summarized in Fig. 3B (red circles). The slope of the fitted linear lines indicates the additional stabilization gained by further glycosylation. Remarkably, both experiments and simulations share the same trend of stabilization as a function of the degree of glycosylation, indicating that the model reproduces not only the general observation of thermal stabilization, but also quantitatively predicts the stabilization effect. The slope of the linear curve for the 63 variants of SH3 is 0.2–0.3, which correlates with a thermal stabilization of 0.6–0.9°C per added glycan. Conjugation with dextran, which is a very long glycan, produced a similar thermal stabilization to that obtained from the attachment of the short lactose glycan (Fig. 3B, red circles). Accordingly, the number of glycans conjugated to the protein governs the degree of stabilization, although the length of the glycans has a minor effect. In our simulations, we also observed the same thermal stabilization by short (5) and long (11) sugar ring glycans (Fig. 3B, filled circles). Some glycosylated SH3 versions, however, are destabilized by the glycans compared with the wild-type SH3. This finding may suggest that stabilization by glycans is sensitive to their location. A similar effect was observed for proteins that are tethered to surfaces, where tethering results in protein stabilization or destabilization depending on the location of the residue used as the tether point (29, 30).
Fig. 3.
Thermal stabilization of the glycosylated variants of the SH3 domain. (A) Specific heat curves of native SH3 (filled line) and of SH3 modified with three (dotted line) and six (dashed line) 11-sugar ring glycans. (B) Effect of glycosylation on protein stability as measured by the change in the Tf. Simulations of 63 SH3 glycosylated variants bearing short glycans comprised 5 sugar rings (open black circles, fitted by a dashed black line; R = 0.805) or long glycans comprised of 11 sugar rings (filled black circles, fitted by a solid black line; R = 0.719). Experimental measurements of stabilization by chemical glycosylation of α-CT (27) with lactose (open red circles, fitted by a dashed red line; R = 0.98) or 10-kDa dextran (filled red circles, fitted by a solid red line; R = 0.936) from the work of Sola et al. (27, 28) are shown. The number of glycans conjugated to α-CT was calculated as the average moles of glycan per mole of α-CT.
The free-energy profiles for the folding of SH3 and two representative variants glycosylated with the long glycan against reaction coordinate Q (at the Tf of the nonglycosylated SH3 domain) are shown in Fig. 4A. At this temperature, the folded state of all three variants has a similar free energy, but the unfolded state is destabilized. The free energy of the unfolded state is larger for the glycosylated than for the nonglycosylated form of SH3. This increase in the free energy of the unfolded state is correlated with the degree of glycosylation and was expected from the increased Tf (the glycans stabilize the protein thermodynamically).
Fig. 4.
Glycosylation effect on the free energy of unfolding. (A) Free-energy profile against the fraction of native contacts (Q used as a reaction coordinate) for native SH3 (filled line) and for SH3 glycosylated with three (dotted line) or six (dashed line) 11-sugar ring glycans. (B) Kinetic effect of glycosylation as measured by the change in the height of the free-energy barrier of unfolding (at the Tf of the native SH3 domain). (C) Free-energy barrier of unfolding at the specific Tf of each glyco-conjugated variant of the SH3 domain (divided by the specific Tf).
Fig. 4B shows a summary of the unfolding free-energy barrier measured at the Tf of the nonglycosylated SH3 domain for all of the studied variants. The energy barrier to unfolding increases with the number of added glycans, implying that glycosylation introduces kinetic stability as well; this effect originates from the thermodynamic stabilization. From the slope of the curve, we can estimate that glycosylation increases the energy barrier by 3.9% per glycan (regardless of the type of glycan attached), compared with the nonglycosylated form of the protein. The energy barrier of each glycosylated variant of SH3 at its specific Tf (divided by the specific kT) is plotted in Fig. 4C. These data are poorly fitted with R = 0.228/0.269 (for short and long glycans, respectively), reflecting a weak correlation between the energy barrier and the number of conjugated glycans. However, it is evident that most of the glycosylated variants have a higher energy barrier than SH3.
The free energy for folding (ΔGf) of all 63 glycosylated SH3 domains is plotted in Fig. 5A. The figure illustrates the thermodynamic stabilization of SH3 conferred by glycosylation. To pinpoint the origin of the thermodynamic stability, we analyzed the free energy of the unfolded and folded states of the 63 variants of glycosylated SH3. In these analyses, the energetic contributions were calculated based on all of the conformations of the ensemble of the folded and unfolded states, weighted by their populations. The free energy of the folded state is not affected by the number of glycans attached (Fig. 5A). However, the free energy of the unfolded state increases as more glycans are added (Fig. 5A), resulting in the overall observed stabilization (i.e., glycosylation increases the free energy of the unfolded state and thereby increases thermodynamic stability). The enthalpic and entropic components of the unfolded and folded states were calculated in a similar way and are shown in Fig. 5 B and C. In the folded state, the entropy is indeed reduced by the glycans, as was speculated above. The reduction in entropy of the folded state, as more glycans are attached, might be an outcome of repressing fluctuations in the folded state and rigidifying the native state. The glycans, therefore, can be viewed as imposing pressure on the protein; as a result, the vibrations of the molecule are suppressed. Furthermore, the folded state of the glycosylated SH3 is characterized by a lower enthalpy than the nonglycosylated variant. The decrease in enthalpy is similar in magnitude to the decrease in entropy. Accordingly, it appears that the decrease in entropy is compensated for by the decrease in enthalpy, and, overall, the free energy of the folded state is unchanged upon glycosylation and does not depend on the degree of glycosylation. By contrast, for the unfolded state, there is an asymmetry in the effect of glycosylation on its entropy and enthalpy. Although both the enthalpy and entropy of the unfolded state increase upon glycosylation, the enthalpy is affected more significantly. This result suggests that the bulky groups of the oligosaccharides restrict the formation of interactions in the unfolded state and force the polypeptide chain to adopt more extended conformations.
Fig. 5.
Effect of glycosylation on the enthalpy (H) and entropy (TS) of the folded and unfolded states. (A) Free energy of folding ΔGf (black) of the SH3 domain and its 63 glycosylated variants as a function of the number of added glycans and the free energy of the folded (Gf, gray) and unfolded (Gun, blue) ensembles. The energetic analysis was performed at the Tf of the nonglycosylated SH3 domain. (B) Enthalpic (gray) and entropic (brown) contributions to the free energy of the folded state ensemble as a function of the number of added glycans. (C) Enthalpic (gray) and entropic (brown) contributions to the free energy of the unfolded state ensemble as a function of the number of added glycans.
To substantiate the assumptions presented above regarding the effect of glycans on the protein in its folded and unfolded states, we calculated the probability of each nonglycosylated SH3 (piSH3) and glycosylated (piglyco) variant residue, i, forming its native contacts during the simulations. We performed these calculations separately for the folded and unfolded ensembles. Fig. 6 presents the differences between the piglyco values of some representative glycosylated SH3 variants and the piSH3 in the folded and unfolded states. The differences are minor in the folded state, indicating that glycosylation does not affect contact formation in the native structure. In the unfolded state, the differences between the glycosylated and nonglycosylated forms are more pronounced. The glycosylated variants are less structured in the unfolded state. Accordingly, the higher enthalpy of the unfolded state of the glycosylated variants with respect to that of the nonglycosylated state is explained by restricting the formation of residual structures in the unfolded state [see also supporting information (SI) Fig. S1 and Fig. S2]. Usually, the structural destabilization of the unfolded state is additive because each glycan introduces a local effect around the glycosylation site. The local structural effect of each glycan is unique to its position and can be treated as a “fingerprint” of each glycosylation site.
Fig. 6.
Effect of glycosylation on the structure of the folded and unfolded state ensembles. (A and B) Structure of the folded (A) and unfolded (B) states measured as the difference between the number of native contacts that are formed by each specific residue of the glycan-conjugated protein and the native SH3 domain. The arrows indicate the positions of the glycosylation sites. The term piSH3 represents the probability of each residue of the nonglycosylated SH3-forming native contacts, whereas piglyco is the equivalent term for the glycosylated variants.
To characterize further the transition state ensemble, we calculated its enthalpy and entropy and observed a reduction in both terms as the number of added glycans increased. The stronger reduction in the entropy yields a higher free-energy barrier when more glycans are attached. Analysis of contact formation in the transition state ensemble of the glycoproteins with respect to the nonglycosylated protein shows that the influence of the glycan is long-ranging, and the glycans increase the number of native contacts formed compared with the number formed by nonglycosylated SH3. This effect correlates with the decreased enthalpy that we observe in the transition state upon increasing the degree of glycosylation.
The thermodynamic analysis demonstrates the uniform entropy of the unfolded state with regard to the addition of glycans. One may expect that the glycans would reduce the entropy of the unfolded state as per the effect of molecular crowding or confinement conditions that limit protein dynamics in the unfolded state. It is very likely that glycosylation reduces the entropy in the unfolded state by confining the conformational space. Nevertheless, the entropy of the unfolded state may be affected by changes in the structural properties of the unfolded state. The smaller residual structure for the glycosylated proteins implies that the polysaccharides unwind the unfolded state and, as a result, increase its configurational entropy. We suggest that these effects cancel out, and therefore there is no significant change in the entropy of the unfolded state upon glycosylation.
Conclusions
In this article, we explored the thermodynamic and kinetic effects of glycosylation on protein folding by using a coarse-grained computational model, and we observed the experimentally reported thermal stabilization of protein upon glycosylation. The thermodynamic stabilization is correlated with the degree of glycosylation and, to a much smaller extent, with the size of the polysaccharides. The stabilization effect of glycosylation is reminiscent of the protein stabilization introduced by molecular crowding or confinement. Although glycosylation at six positions on SH3 shifted the Tf by ≈1%, molecular crowding may yield a change of ≈7% in the Tf (31), and a highly confined space may shift the Tf by ≈25% (32–35). It is likely that glycoproteins in a crowded or confined environment will exhibit an enhanced stabilization effect, yet the additivity of glycans and crowding agents or confinements needs to be thoroughly investigated in the future.
The stabilization effect depends on the position of the glycans; thus, the same degree of glycosylation may result in a different thermal effect depending on the location of the sugars. Importantly, thermodynamic stabilization is accompanied by kinetic stabilization, with the unfolding barrier increased by ≈20% for the highly glycosylated variants. A thorough thermodynamic analysis revealed that the main origin for the stabilization introduced by glycosylation is the unwinding of the residual native structure formed in the unfolded state, which results in a higher enthalpy. This effect serves as another indication of the importance of the unfolded state in controlling and shifting the thermodynamics and kinetics of protein folding (36–38).
The coarse-grained model used here reproduces the known experimental thermal stabilizing effect of glycans. In the current model, the glycans are modeled as entropic polymers that do not form any specific stabilizing interactions with the protein chain. Accordingly, we suggest that the glycan effect lies in modulating the entropy and the residual structure of the unfolded state. Yet, supplementing the model with other features of glycans may allow other questions to be addressed. Modeling non-native interactions may allow exploring structural changes due to glycosylation as often found in phosphorylation (39, 40). Favorable interactions between the carbohydrates and the protein in both the folded and unfolded states are possible and are seen in some crystal structures. Although in most cases these interactions do not contribute to the stability of the protein, specific cases also were reported where these interactions were crucial to protein stability. Furthermore, it is essential to model the solvent effect in more detail to include the solvent entropy, in addition to the configurational entropy, of the protein.
Our article suggests that glycosylation can enrich and modulate the biophysical properties of proteins and offers an alternative way to design thermally stabilized proteins. This stabilization strategy may aid in designing other polymer-conjugated proteins, such as PEGylated proteins (i.e., proteins conjugated to polyethylene glycol), which may share features similar to those of sugar-conjugated proteins.
Materials and Methods
In this article, we applied a native topology-based model (Go model) to study the folding of glycoproteins. In this model, each amino acid and each sugar ring is represented by a single bead centered at the Cα and C1 atoms, respectively. All secondary and tertiary native contacts between amino acids are represented by the Lennard–Jones potential without any discrimination between the various chemical types of the interactions. The Hamiltonian of the system and its parameters can be found elsewhere (6).
The protein is introduced by five terms (for bonds, angles, torsion angles, the Lennard–Jones term for native interactions, and the excluded volume term for non-native interactions) that define the properties of its folding. The glycan conjugate is introduced by fewer terms, namely, a bond term that defines its connectivity and by an excluded volume term that introduces the large volume of each sugar ring. The excluded volume distances between a pair of protein beads and between an amino acid and sugar bead are 4 and 6 Å, respectively. We neglect possible interactions between the glycans and the proteins, as well as between the glycans. The rigidity of the glycans is introduced by including an angle potential for the native angles between the beads that represent the sugar rings.
Using this model, multiple trajectories simulated using the Langevin equation (γ = 0.01) were collected to look for numerous unfolding/folding transitions at various temperatures. The attached glycans introduce inherent inhomogeneity in the degrees of freedom of the system (41), which requires special care in the choice of the simulation thermostat. Recently (41), we showed that the Berendsen thermostat failed in dealing with such systems, whereas the Langevin thermostat successfully maintained the temperature of the simulation. The trajectories were analyzed by using the weighted histogram analysis method (42) to study the folding thermodynamics. Comparing between the calculated thermodynamic features of proteins of different sizes was problematic because the energy of the native structure depended on the size of the protein. To overcome this obstacle, we used only the energy term for the native contacts, which was identical in all of the glycosylated variants and corresponded only to the SH3 protein moiety without the conjugated glycans. The native nonbonded contacts and their distances were calculated by using contact of structural units (43). The structures of the SH3 domain glycol-conjugated variants were built by using the Glyprot web server (44).
Water was not explicitly represented in our calculations, yet the water molecules that were elemental for maintaining the structure were indirectly represented in native topology-based models that ensured correct folding. Moreover, the random forces applied by the water molecules on the solute were introduced by the Langevin dynamics in the simulations. Additionally, the water may have affected the thermodynamics of biomolecules through the formation of hydrogen bonds with the protein backbone and with some side chains. In calculating the thermodynamic properties of the protein, the water also was pivotal due to the often large solvation entropy that is released upon folding. In the current article, we treated the glycosylation as a perturbation to the protein and assumed that the solvation entropy was identical for the glycosylated and nonglycosylated proteins. Accordingly, in this article, we focused on the effect of glycosylation on the configuration entropy.
Supplementary Material
Acknowledgments.
We thank Nathan Sharon for guiding us in the fascinating world of glycoproteins and for many suggestions. This work was supported, in part, by the Kimmelman Center for Macromolecular Assemblies, the Clore Center for Biological Physics, the Center for Complexity Science (to Y.L.), and the Clore Foundation (to D.S.-B). Y.L. is the incumbent of the Lilian and George Lyttle Career Development Chair.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801340105/DCSupplemental.
References
- 1.Fersht AR, Daggett V. Protein folding and unfolding at atomic resolution. Cell. 2002;108:573–582. doi: 10.1016/s0092-8674(02)00620-7. [DOI] [PubMed] [Google Scholar]
- 2.Oliveberg M, Wolynes PG. The experimental survey of protein-folding energy landscapes. Q Rev Biophys. 2005;38:245–288. doi: 10.1017/S0033583506004185. [DOI] [PubMed] [Google Scholar]
- 3.Onuchic JN, Wolynes PG. Theory of protein folding. Curr Opin Struct Biol. 2004;14:70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Papoian GA, Ulander J, Eastwood MP, Luthey-Schulten Z, Wolynes PG. Water in protein structure prediction. Proc Natl Acad Sci USA. 2004;101:3352–3357. doi: 10.1073/pnas.0307851100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin W, Kambara O, Sasakawa H, Tamura A, Takada S. De novo design of foldable proteins with smooth folding funnel: Automated negative design and experimental verification. Structure. 2003;11:581–590. doi: 10.1016/s0969-2126(03)00075-3. [DOI] [PubMed] [Google Scholar]
- 6.Clementi C, Nymeyer H, Onuchic JN. Topological and energetic factors: What determines the structural details of the transition state ensemble and “en-route” intermediates for protein folding? An investigation for small globular proteins. J Mol Biol. 2000;298:937–953. doi: 10.1006/jmbi.2000.3693. [DOI] [PubMed] [Google Scholar]
- 7.Varki A. Biological roles of oligosaccharides: All of the theories are correct. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lis H, Sharon N. Protein glycosylation: Structural and functional aspects. Eur J Biochem. 1993;218:1–27. doi: 10.1111/j.1432-1033.1993.tb18347.x. [DOI] [PubMed] [Google Scholar]
- 9.Spiro RG. Protein glycosylation: Nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2002;12:43R–56R. doi: 10.1093/glycob/12.4.43r. [DOI] [PubMed] [Google Scholar]
- 10.Dwek RA. Glycobiology: Toward understanding the function of sugars. Chem Rev. 1996;96:683–720. doi: 10.1021/cr940283b. [DOI] [PubMed] [Google Scholar]
- 11.Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 12.Parodi AJ. Role of N-oligosaccharide endoplasmic reticulum processing reactions in glycoprotein folding and degradation. Biochem J. 2000;348(Pt 1):1–13. [PMC free article] [PubMed] [Google Scholar]
- 13.Mitra N, Sinha S, Ramya TN, Surolia A. N-linked oligosaccharides as outfitters for glycoprotein folding, form and function. Trends Biochem Sci. 2006;31:156–163. doi: 10.1016/j.tibs.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Mitra N, Sharon N, Surolia A. Role of N-linked glycan in the unfolding pathway of Erythrina corallodendron lectin. Biochemistry. 2003;42:12208–12216. doi: 10.1021/bi035169e. [DOI] [PubMed] [Google Scholar]
- 15.Wyss DF, et al. Conformation and function of the N-linked glycan in the adhesion domain of human CD2. Science. 1995;269:1273–1278. doi: 10.1126/science.7544493. [DOI] [PubMed] [Google Scholar]
- 16.Riederer MA, Hinnen A. Removal of N-glycosylation sites of the yeast acid phosphatase severely affects protein folding. J Bacteriol. 1991;173:3539–3546. doi: 10.1128/jb.173.11.3539-3546.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudd PM, et al. Glycoforms modify the dynamic stability and functional activity of an enzyme. Biochemistry. 1994;33:17–22. doi: 10.1021/bi00167a003. [DOI] [PubMed] [Google Scholar]
- 18.Wang C, Eufemi M, Turano C, Giartosio A. Influence of the carbohydrate moiety on the stability of glycoproteins. Biochemistry. 1996;35:7299–7307. doi: 10.1021/bi9517704. [DOI] [PubMed] [Google Scholar]
- 19.Sinha S, Surolia A. Attributes of glycosylation in the establishment of the unfolding pathway of soybean agglutinin. Biophys J. 2007;92:208–216. doi: 10.1529/biophysj.106.092668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sola RJ, Griebenow K. Chemical glycosylation: New insights on the interrelation between protein structural mobility, thermodynamic stability, and catalysis. FEBS Lett. 2006;580:1685–1690. doi: 10.1016/j.febslet.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 21.Hansia P, Dev S, Surolia A, Vishveshwara S. Insight into the early stages of thermal unfolding of peanut agglutinin by molecular dynamics simulations. Proteins. 2007;69:32–42. doi: 10.1002/prot.21512. [DOI] [PubMed] [Google Scholar]
- 22.Bosques CJ, Tschampel SM, Woods RJ, Imperiali B. Effects of glycosylation on peptide conformation: A synergistic experimental and computational study. J Am Chem Soc. 2004;126:8421–8425. doi: 10.1021/ja049266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandal TK, Mukhopadhyay C. Effect of glycosylation on structure and dynamics of MHC class I glycoprotein: A molecular dynamics study. Biopolymers. 2001;59:11–23. doi: 10.1002/1097-0282(200107)59:1<11::AID-BIP1001>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 24.Petrescu AJ, Milac AL, Petrescu SM, Dwek RA, Wormald MR. Statistical analysis of the protein environment of N-glycosylation sites: Implications for occupancy, structure, and folding. Glycobiology. 2004;14:103–114. doi: 10.1093/glycob/cwh008. [DOI] [PubMed] [Google Scholar]
- 25.Wyss DF, Wagner G. The structural role of sugars in glycoproteins. Curr Opin Biotechnol. 1996;7:409–416. doi: 10.1016/s0958-1669(96)80116-9. [DOI] [PubMed] [Google Scholar]
- 26.DeKoster GT, Robertson AD. Thermodynamics of unfolding for Kazal-type serine protease inhibitors: Entropic stabilization of ovomucoid first domain by glycosylation. Biochemistry. 1997;36:2323–2331. doi: 10.1021/bi962580b. [DOI] [PubMed] [Google Scholar]
- 27.Sola RJ, Al-Azzam W, Griebenow K. Engineering of protein thermodynamic, kinetic, and colloidal stability: Chemical glycosylation with monofunctionally activated glycans. Biotechnol Bioeng. 2006;94:1072–1079. doi: 10.1002/bit.20933. [DOI] [PubMed] [Google Scholar]
- 28.Sola RJ, Rodriguez-Martinez JA, Griebenow K. Modulation of protein biophysical properties by chemical glycosylation: Biochemical insights and biomedical implications. Cell Mol Life Sci. 2007;64:2133–2152. doi: 10.1007/s00018-007-6551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedel M, Baumketner A, Shea JE. Stability of a protein tethered to a surface. J Chem Phys. 2007;126 doi: 10.1063/1.2464114. 095101. [DOI] [PubMed] [Google Scholar]
- 30.Friedel M, Baumketner A, Shea JE. Effects of surface tethering on protein folding mechanisms. Proc Natl Acad Sci USA. 2006;103:8396–8401. doi: 10.1073/pnas.0601210103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheung MS, Klimov D, Thirumalai D. Molecular crowding enhances native state stability and refolding rates of globular proteins. Proc Natl Acad Sci USA. 2005;102:4753–4758. doi: 10.1073/pnas.0409630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thirumalai D, Lorimer GH. Chaperonin-mediated protein folding. Annu Rev Biophys Biomol Struct. 2001;30:245–269. doi: 10.1146/annurev.biophys.30.1.245. [DOI] [PubMed] [Google Scholar]
- 33.Takagi F, Koga N, Takada S. How protein thermodynamics and folding mechanisms are altered by the chaperonin cage: Molecular simulations. Proc Natl Acad Sci USA. 2003;100:11367–11372. doi: 10.1073/pnas.1831920100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu WX, Wang J, Wang W. Folding behavior of chaperonin-mediated substrate protein. Proteins. 2005;61:777–794. doi: 10.1002/prot.20689. [DOI] [PubMed] [Google Scholar]
- 35.Friedel M, Sheeler DJ, Shea JE. Effects of confinement and crowding on the thermodynamics and kinetics of folding of a minimalist beta-barrel protein. J Chem Phys. 2003;118:8106–8113. [Google Scholar]
- 36.Shortle D, Ackerman MS. Persistence of native-like topology in a denatured protein in 8 M urea. Science. 2001;293:487–489. doi: 10.1126/science.1060438. [DOI] [PubMed] [Google Scholar]
- 37.Pappu RV, Srinivasan R, Rose GD. The Flory isolated-pair hypothesis is not valid for polypeptide chains: Implications for protein folding. Proc Natl Acad Sci USA. 2000;97:12565–12570. doi: 10.1073/pnas.97.23.12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho JH, Sato S, Horng JC, Anil B, Raleigh DP. Electrostatic interactions in the denatured state ensemble: Their effect upon protein folding and protein stability. Arch Biochem Biophys. 2008;469:20–28. doi: 10.1016/j.abb.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen T, Zong C, Hamelberg D, McCammon JA, Wolynes PG. The folding energy landscape and phosphorylation: Modeling the conformational switch of the NFAT regulatory domain. FASEB J. 2005;19:1389–1395. doi: 10.1096/fj.04-3590hyp. [DOI] [PubMed] [Google Scholar]
- 40.Latzer J, Shen T, Wolynes PG. Conformational switching upon phosphorylation: A predictive framework based on energy landscape principles. Biochemistry. 2008;47:2110–2122. doi: 10.1021/bi701350v. [DOI] [PubMed] [Google Scholar]
- 41.Mor A, Ziv G, Levy Y. Simulations of proteins with inhomogeneous degrees of freedom: The effect of thermostats. J Comp Chem. 2008 doi: 10.1002/jcc.20951. in press. [DOI] [PubMed] [Google Scholar]
- 42.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The weighted histogram analysis method for free-energy calculations on biomolecules. 1. The Method. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 43.Sobolev V, Wade RC, Vriend G, Edelman M. Molecular docking using surface complementarity. Proteins. 1996;25:120–129. doi: 10.1002/(SICI)1097-0134(199605)25:1<120::AID-PROT10>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 44.Bohne-Lang A, von der Lieth CW. GlyProt: In silico glycosylation of proteins. Nucleic Acids Res. 2005;33:W214–W219. doi: 10.1093/nar/gki385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.