Abstract
The p110 isoforms of phosphoinositide 3-kinase (PI3K) are acutely regulated by extracellular stimuli. The class IA PI3K catalytic subunits (p110α, p110β, and p110δ) occur in complex with a Src homology 2 (SH2) domain-containing p85 regulatory subunit, which has been shown to link p110α and p110δ to Tyr kinase signaling pathways. The p84/p101 regulatory subunits of the p110γ class IB PI3K lack SH2 domains and instead couple p110γ to G protein-coupled receptors (GPCRs). Here, we show, using small-molecule inhibitors with selectivity for p110β and cells derived from a p110β-deficient mouse line, that p110β is not a major effector of Tyr kinase signaling but couples to GPCRs. In macrophages, both p110β and p110γ contributed to Akt activation induced by the GPCR agonist complement 5a, but not by the Tyr kinase ligand colony-stimulating factor-1. In fibroblasts, which express p110β but not p110γ, p110β mediated Akt activation by the GPCR ligands stromal cell-derived factor, sphingosine-1-phosphate, and lysophosphatidic acid but not by the Tyr kinase ligands PDGF, insulin, and insulin-like growth factor 1. Introduction of p110γ in these cells reduced the contribution of p110β to GPCR signaling. Taken together, these data show that p110β and p110γ can couple redundantly to the same GPCR agonists. p110β, which shows a much broader tissue distribution than the leukocyte-restricted p110γ, could thus provide a conduit for GPCR-linked PI3K signaling in the many cell types where p110γ expression is low or absent.
Keywords: gene targeting, signaling, tyrosine kinase, Akt, insulin
The lipid second messengers generated by phosphoinositide 3-kinases (PI3Ks) regulate a wide variety of cellular functions such as cell growth, proliferation, differentiation, and survival and have been implicated in cancer, inflammation, and diabetes. Mammals have eight isoforms of PI3K, which have been divided in three classes (1). Thus far, attention has focused mainly on the class I PI3Ks that are acutely activated by extracellular ligands. These heterodimers consist of a p110 catalytic subunit in complex with a regulatory subunit and have further been subdivided into class IA and IB PI3Ks. The class IA catalytic subunits (p110α, p110β, and p110δ) are in complex with an Src homology 2 (SH2) domain-containing regulatory subunit (of which there are five species, often referred to as p85s) that binds phosphoTyr in intracellular proteins, allowing recruitment of p85/p110 complexes to the membrane. The class IB regulatory subunits (p84, p101) do not have SH2 domains and link the single class IB PI3K catalytic subunit (p110γ) to G protein-coupled receptors (GPCRs).
Over the last few years, the cellular signaling contexts and physiological roles of p110α, p110δ, and p110γ have become clearer, because of the generation of gene-targeted mice and small-molecule inhibitors for these PI3K isoforms. In contrast, very little is known about p110β, both at the cellular and organismal level. One confounding factor has been the very early embryonic lethality of the p110β KO mice and the fact that proliferating cells could not be derived from these embryos (2). Recent progress has been made by the generation of small-molecule inhibitors with selectivity for p110β, allowing scientists to define a role for p110β in platelet function and thrombus formation (3). Some evidence has also been presented for the coupling of p110β to GPCRs, either by in vitro studies that documented activation of p110β by Gβγ subunits (4, 5) or in cellular experiments where p110β function was probed by microinjection of neutralizing antibodies to p110β (6, 7), RNAi against p110β (8), or overexpression of p110β (8, 9). Here, we have used pharmacological tools with selectivity for p110β (3), in conjunction with cells derived from a mouse line in which p110β has been inactivated by gene targeting, to investigate the role of p110β in PI3K signaling downstream of Tyr kinase and GPCR ligands.
Results
Conditional Genetic Inactivation of p110β.
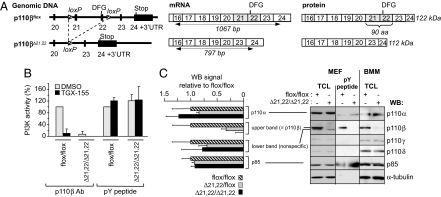
We have created a mouse line in which exons 21 and 22 of the kinase domain of p110β are flanked by loxP sites [supporting information (SI) Fig. S1]. This floxed p110β allele, which is further referred to as p110βflox, is schematically shown in Fig. 1A. Exon 22 contains the DFG motif and the activation loop, which are critical for the activity of kinases. The loxP sites were positioned in such a way that, after treatment with Cre, the locus is expected to give rise to an mRNA in which exon 20 is spliced in-frame onto exon 23, encoding an internally truncated p110β protein with a predicted Mr of ≈112 kDa (further referred to as p110βΔ21,22) instead of the ≈122-kDa WT protein.
Fig. 1.
Genetic inactivation of p110β in mice. (A) Schematic representation of genomic DNA, mRNA, and protein primary structure of the p110βflox and p110βΔ21,22 locus. Exons 22 and 23 of Pik3cb were flanked by loxP sites as described in Fig. S1, creating a floxed p110β allele that is further referred to as p110βflox. Exon 22 encodes the DFG domain and activation loop that are critical for p110β kinase activity. Constitutive deletion of exons 21 and 22 in mice was achieved by intercrossing p110βflox/flox mice with Cre deleter mice, creating the p110βΔ21,22 allele. The loxP sites were positioned in such a way that, after treatment with Cre, the locus is expected to give rise to mRNA in which exon 20 is spliced in-frame onto exon 23, now encoding an internally truncated p110β protein lacking amino acids 886–975 in the catalytic domain. This p110βΔ21,22 protein is predicted to be kinase-inactive, with a ≈10-kDa reduction in Mr relative to the WT p110β protein and to retain reactivity with antisera raised against the extreme C terminus of p110β. (Left) Exon sequences are represented by filled black rectangles, and intron sequences are indicated by a horizontal black line. The loxP sites are shown as hatched triangles with the pointed end indicating orientation. (Center) Exon boundaries are represented from exons 16 to 24. The positions of the primers used for PCR screening are designated by arrows together with the expected amplification products and their size (bp). (Right) Schematic representation and predicted Mr of WT and mutant p110β proteins, based on exon boundaries from exons 16 to 24. (B) Effect of p110β deletion on in vitro lipid kinase activity. Homogenates of the indicated MEFs were immunoprecipitated by using p110β Abs or absorbed onto PDGF receptor phosphoTyr peptide (pY peptide) immobilized to Sepharose (which binds all class IA PI3K regulatory subunits), followed by in vitro lipid kinase assay with or without 100 nM TGX-155. The level of p110β and p85 in the indicated cell fractions was verified by immunoblotting (data not shown). (C) Effect of p110β deletion on PI3K isoform expression. (Left) Analysis of p110β protein expression in MEFs from p110βflox/flox, p110βflox/Δ21,22, and p110βΔ21,22/Δ21,22 mice by Western blotting. Immunoblot signal intensities obtained with Abs to p110α, p110β, and p85 were expressed relative to signals obtained with antibodies to α-tubulin for at least three different embryos and expressed relative to the signals in p110βflox/flox MEFs. Quantification of the signals detected by the sc-602 p110β Abs revealed that only the intensity of the upper band is decreased upon Cre treatment, whereas the intensity of the lower band is unaffected. p110α expression was not affected in each genotype. p110γ and p110δ were hardly detectable in fibroblasts and could not be reliably quantified. (Right) Total cell lysates or pY-peptide pull-downs were immunoblotted by using the indicated Abs. In NIH 3T3 cells and MEFs (but not in BMMs), the anti-p110β Abs used for Western blotting (sc-602) recognize a nonspecific protein just below the specific p110β signal, indicated as lower band (aspecific) and upper band (p110β), respectively. It is only the upper band that disappears upon p110β deletion.
From crosses with heterozygous parents, we obtained ≈30% of expected numbers of viable mice, homozygous for the p110βΔ21,22 allele (p110βΔ21,22/Δ21,22 mice), indicating lethality with incomplete penetrance, for reasons that are unclear at this point of time. Mouse embryonic fibroblasts (MEFs) and bone marrow-derived macrophages (BMMs) from viable p110βΔ21,22/Δ21,22 mice were used for further study. RT-PCR and DNA sequencing on mRNA prepared from p110βΔ21,22/Δ21,22 MEFs showed that the mutant p110β locus gave rise to the expected truncated mRNA (Fig. S1I and data not shown).
In control p110βflox/flox MEFs, lipid kinase activity in p110β immunoprecipitates, made using Abs against the C terminus of p110β, was sensitive to the p110β inhibitor TGX-155 (Fig. 1B). In contrast, no significant lipid kinase activity could be recovered in p110β immunoprecipitates (IPs) from p110βΔ21,22/Δ21,22 MEFs (Fig. 1B). PI3K activity associated with a phosphoTyr peptide matrix (which binds all p85 species; referred to as pY peptide) was unaffected in p110βΔ21,22/Δ21,22 MEFs (Fig. 1B), indicating that p110β contributes minimally to p85-associated PI3K activity in unstimulated fibroblasts.
Transient overexpression in HEK293 cells of the p110βΔ21,22 cDNA cloned from p110βΔ21,22 MEFs revealed that the p110βΔ21,22 protein could be translated from this mRNA (Fig. S1K). As expected, the ≈122-kDa WT p110β protein could no longer be detected in p110βΔ21,22/Δ21,22 MEFs and BMMs upon immunoblotting with C-terminal (Fig. 1C Right) or N-terminal antibodies to p110β (Fig. S1J). However, no evidence for the ≈112-kDa p110βΔ21,22 protein could be found (Fig. 1C and Fig. S1J), indicating that this truncated protein is either not produced, is further truncated, or is unstable [note that the lower Mr signal observed with the p110β Cter Abs in MEFs is a nonspecific signal that is not altered in p110βΔ21,22/Δ21,22 cells (Fig. 1C); this lower MW signal is not observed in BMMs]. The levels (Fig. 1C) and activity (Fig. S1L) of p110α were unaffected. The observation that expression of the p85 regulatory subunit and the other PI3K isoforms is not affected in MEFs and BMMs (Fig. 1C) argues against the degradation of the entire p110β protein, which would most likely lead to a reduction in p85 levels (10). Thus, in p110βΔ21,22/Δ21,22 cells, p110β activity is absent, yet the p85/p110 stoichiometry is unaltered.
p110β and p110γ Signal Downstream of the Same GPCR Ligands in Cells Expressing All Class I PI3Ks.
We first assessed the role of p110β in early PI3K signaling in BMMs that express all class I p110 isoforms, including p110γ (Fig. 1C).
Phosphorylation of Akt and Erk induced by the Tyr kinase ligand colony-stimulating factor-1 (CSF-1) was not affected by pharmacological or genetic inactivation of p110β or p110γ (Fig. S2 A-C; p110γ was inhibited by using AS604850, a small-molecule inhibitor with selectivity for p110γ (11)). These data indicate that p110β and p110γ do not play a major role in Tyr kinase-induced PI3K activation in BMMs. This was previously reported for p110γ (11–13), but is somewhat unexpected for p110β, given that injection of neutralizing Abs to p110β has been shown to block CSF-1-induced chemotaxis in a macrophage cell line (14).
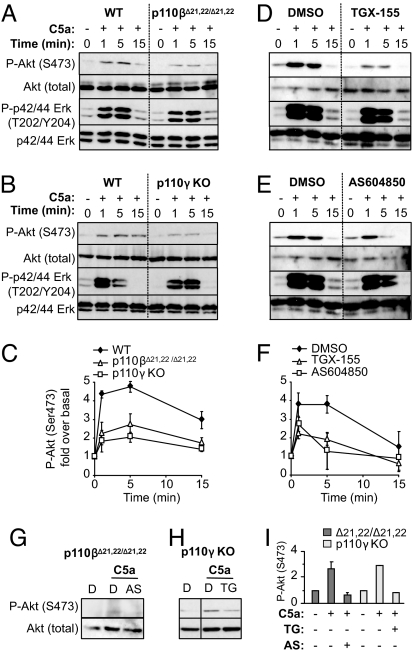
In contrast to Tyr kinase ligands, Akt phosphorylation induced by the GPCR agonist complement 5a (C5a) was substantially, but not completely, inhibited by inactivation of p110β, either genetically (Fig. 2 A and C) or pharmacologically (Fig. 2 D and F). The residual C5a-induced Akt phosphorylation in the absence of p110β activity could be blocked by the p110γ inhibitor AS604850 (Fig. 2 G and I), indicating that p110γ controlled this component of the signal.
Fig. 2.
Role of p110β and p110γ in cell signaling in macrophages. (A and B) Starved BMMs of the indicated genotype were stimulated with C5a, followed by immunoblotting using the indicated Abs. (D and E) Starved WT BMMs were treated for 30 min with TGX-155 (0.5 μM) (D) or AS604850 (1 μM) (E), followed by C5a stimulation and immunoblotting using the indicated Abs. (C and F) Quantification of at least two independent experiments was performed, and data are shown as fold over P-Akt under unstimulated conditions. (G–I) Starved BMMs of the indicated genotype were treated for 30 min with AS604850 (1 μM) or TGX-155 (0.5 μM) and stimulated for 5 min with C5a, followed by immunoblotting using the indicated Abs. I shows the quantification of the experiments in G and H. Data are expressed as fold over P-Akt under unstimulated conditions.
In line with previous reports (12), C5a-induced Akt phosphorylation was decreased in p110γ KO BMMs (Fig. 2 B and C) and in WT BMMs upon treatment with AS604850 (Fig. 2 E and F). However, residual C5a-stimulated Akt phosphorylation was also observed under those conditions (see quantification of Western blot signal in Fig. 2 C and F). This activity appears to be mediated by p110β, because TGX-155 treatment inhibited the residual p-Akt signal in C5a-stimulated p110γ KO BMMs (Fig. 2 H and I).
Pertussis toxin (PTx) treatment blocked the C5a-induced pAkt signal in p110γ KO and p110βΔ21,22/Δ21,22 cells (Fig. S2 D and E), indicating that C5a-stimulated Akt phosphorylation mediated by p110β and p110γ, respectively, is G protein-dependent.
Taken together, these data show that p110β signals effectively downstream of a GPCR in cells and that p110β and p110γ are activated downstream of the same GPCR ligand to mediate Akt phosphorylation. Genetic or pharmacological inactivation of p110β or p110γ did not affect CSF-1 or C5a-induced Erk phosphorylation (Fig. 2 A, B, D, and E).
p110β Controls GPCR-Induced Early PI3K Signaling in Fibroblasts.
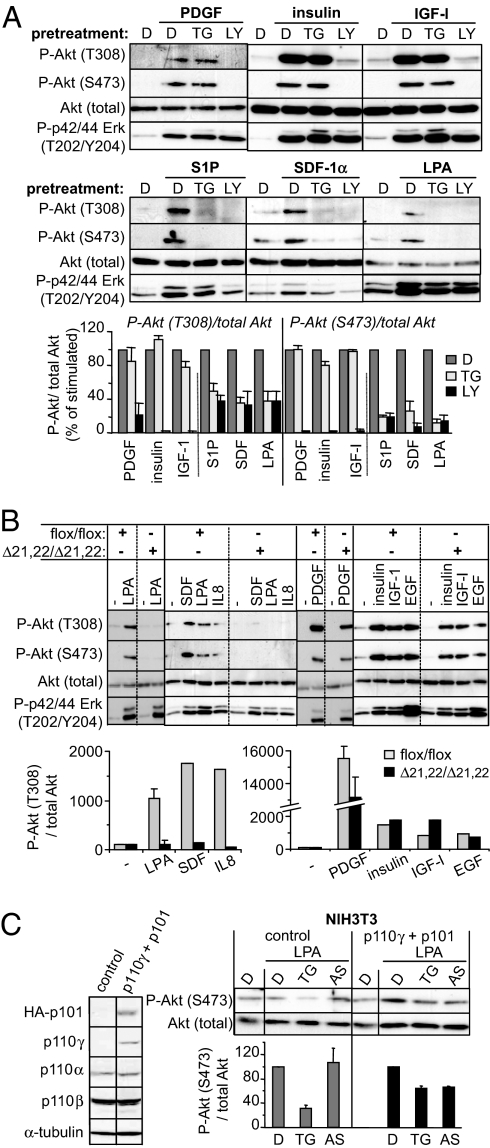
We next tested the role of p110β in Tyr kinase and GPCR signaling in NIH 3T3 and MEFs, which mainly express p110α and p110β and have undetectable or low levels of p110γ and p110δ, respectively (Fig. 1C and Fig. S3A). Absolute quantification of class IA p110 isoforms in NIH 3T3 cells showed that p110β is expressed at much higher levels than p110α and p110δ [12,000 versus 2,000 and 1,000 molecules per cell, respectively (15)]. However, given that barely detectable levels of p110γ can have a major biological impact [such as in cardiomyocytes (16)], we first tested the potential role of p110γ in these cells. The p110γ inhibitor AS604850 did not impair lysophosphatidic acid (LPA)-induced phosphorylation of Akt in NIH 3T3 cells (Fig. S3B). Moreover, in p110γ KO MEFs, GPCR stimuli such as sphingosine-1-phosphate (S1P) and LPA could still induce Akt phosphorylation (Fig. S3C). This Akt phosphorylation was sensitive to TGX-155 but not to AS604850 (Fig. S3C), suggesting that p110β is the main GPCR-coupled PI3K isoform in fibroblasts.
To further substantiate these findings, a broader range of agonists was tested in NIH 3T3 cells. Akt phosphorylation induced by PDGF, insulin, or insulin-like growth factor 1 (IGF1) was sensitive to LY294002 but was not blocked by TGX-155 (Fig. 3A Top). This finding is in stark contrast to Akt phosphorylation induced by each of the GPCR ligands tested [S1P, stromal cell-derived factor (SDF-1α), LPA], which were equally sensitive to TGX-155 and LY294002 in each case (Fig. 3A Middle; for quantification of the results of three independent experiments with all six stimuli, see Fig. 3A Bottom). Similar results were obtained in WT MEFs upon stimulation with PDGF or GPCR ligands such as S1P and LPA (Fig. S3D) and upon using TGX-115 and TGX-221, two alternative inhibitors from the same chemical series as TGX-155 (3) (data not shown).
Fig. 3.
Role of p110β and p110γ in cell signaling in fibroblasts. (A) (Top and Middle) Starved NIH 3T3 cells were treated for 1 h with TGX-155 (TG; 1 μM), LY294002 (LY; 20 or 5 μM), or vehicle DMSO (D) followed by stimulation for 10 min with the indicated ligands. Total cell lysates were immunoblotted with the indicated Abs. A representative immunoblot is shown. (Bottom) Quantification of three independent experiments was performed, and data are presented as percentage of ligand-induced P-Akt/total Akt. (B) Starved MEFs of indicated genotype were stimulated with indicated stimuli for 10 min and immunoblotted with the indicated Abs. A representative experiment performed with MEFs isolated from different embryos is shown. (C) p110 isoform expression in NIH 3T3 cells transfected with empty vector (NIH 3T3 control) or p110γ and its regulatory subunit HA-p101 (p110γ + p101). Starved cells were treated for 1 h with TGX-155 (1 μM) or AS604850 (1 μM), followed by stimulation for 10 min with LPA and immunoblotting with the indicated Abs.
LPA-induced, but not PDGF-induced, phosphorylation of Akt was also inhibited by TGX-155 when tested at different time points (Fig. S3E). TGX-155 inhibited LPA-induced T308 and S473 phosphorylation of Akt in a dose-dependent manner, with an IC50 of 0.1 μM (Fig. S3F), in line with the PI3K isoform specificity of this compound (see Materials and Methods). Occasionally, an increase in LPA-induced S473 phosphorylation of Akt was observed at very low doses of TGX-155 (0.01 μM; data not shown) for reasons that are unclear. PTx pretreatment decreased LPA-induced Akt phosphorylation in NIH 3T3 cells, but had no effect on PDGF-induced Akt phosphorylation (Fig. S3G).
In p110βΔ21,22/Δ21,22 MEFs, Tyr kinase (PDGF, insulin, IGF1, EGF)-induced Akt phosphorylation was largely unaffected, in contrast to Akt phosphorylation induced by the GPCR ligands LPA, SDF-1, and IL-8, which was almost completely blocked (Fig. 3B). Similar results were obtained with MEFs derived from mice that are homozygous for a kinase-dead allele of p110β (p110βD931A/D931A; data not shown). Also acute genetic deletion of p110β [by 4-hydroxytamoxifen treatment of MEFs from p110βflox/flox mice crossed onto the ROSA-CreERT2 mice (17), resulting in activation of Cre by its translocation from the cytosol to the nucleus], reduced LPA-induced Akt phosphorylation, with the residual pAkt signal being no longer sensitive to TGX-155 (Fig. S3H).
Taken together, these data show that p110β is critical for early PI3K signaling induced by GPCRs but is not a major transducer downstream of receptor Tyr kinases.
p110γ Can Complement p110β Function in GPCR-Induced PI3K Signaling in Fibroblasts.
To test whether p110γ could in principle signal downstream of GPCRs in fibroblasts, we stably transfected p110γ, together with its p101 regulatory subunit, into NIH 3T3 cells (Fig. 3C Left). In control transfectants, the p110γ inhibitor AS604850 did not decrease LPA-induced Akt phosphorylation (Fig. 3C). In contrast, NIH 3T3 cells expressing p110γ became sensitive to AS604850 and also showed reduced sensitivity to the p110β inhibitor TGX-155 (Fig. 3C). These data further substantiate the notion that p110β and p110γ can signal in the same GPCR signaling pathways.
Discussion
Here, we define a key role for p110β in early PI3K signaling downstream of GPCRs. This is not only the case in cells that do not express p110γ, the other well established GPCR-linked PI3K, but also in leukocytes that coexpress p110β and p110γ. Our data show that in the latter cells p110β acts in concert with p110γ to provide full PI3K activity downstream of GPCRs, contradicting previous reports that indicated that p110γ inactivation leads to full blockade of Akt phosphorylation by GPCR ligands in macrophages (11–13) but also in other leukocytes such as neutrophils (18) or mast cells (19). Further studies using a broader range of ligands and cell types will be essential to determine the relative importance of p110β and p110γ in signaling and biological outputs.
Somewhat unexpectedly, our data show that p110β does not contribute substantially to Akt activation by Tyr kinase ligands. This finding is surprising, given that the absolute amount of p110β in the cell types studied here is three to six times higher than that of p110α (15). However, our observations are in line with recent reports that p110α is the major PI3K isoform in Tyr kinase signaling by insulin and IGF1, with a minimal contribution of p110β (20, 21). Also, p110β has been found to be recruited to the PDGF receptor, without contributing to Akt activation and the biological functions induced by PDGF (7, 22, 23). Key differences between p110α and p110β in insulin signaling include selective recruitment of p110α over p110β to IRS-1/2 complexes (20) and the lack of p110β lipid kinase activity in these complexes (20, 21). It is possible that p110β is precluded from effective recruitment to membrane-bound receptors because of its association with the Rab5 small GTPase (24), which is found mainly on endosomes. Parameters that could influence the low activity of p110β in Tyr kinase complexes include the low specific activity of p110β compared with p110α (25) and the notion that full activation of p110β upon Tyr-mediated recruitment may require the presence of Gβγ subunits (4, 5, 8, 26). p110β could thus become activated by Tyr kinase receptors under conditions whereby cells receive parallel stimulation through GPCRs, a scenario that may very well be operational in vivo when cells are confronted with a multitude of stimuli. For example, this could occur upon direct activation of Src-family Tyr kinases by GPCRs (27).
Our studies do not exclude a role for p85-mediated recruitment of p110β, given that our screen of Tyr kinase ligands and cell types, as well as kinetics and dose of stimulation, has not been exhaustive. Indeed, other stimuli and biological responses that activate Tyr kinases may engage p110β, including apoptotic cell and FcγR-mediated phagocytosis and CSF-1-stimulated chemotaxis in macrophages (28), EGF-induced DNA synthesis in breast cancer cells (29), FcεRI-activated calcium influx in mast cells (30), and insulin signaling in endothelial and hepatic cell lines (22, 31). It is possible that p110β, while not being a major effector in early PI3K signaling, could contribute to Tyr kinase-driven PI3K signaling at later time points and in different signaling contexts and/or modulate signaling through other PI3K isoforms. Evidence for the latter has been documented in insulin signaling, whereby “basal” p110β activity seems to set the threshold for activation of p110α (21, 31). It is tempting to speculate that p110β-mediated GPCR-PI3K signaling through serum components (such as LPA) in the cell models used in these studies contributes to this basal PI3K activity.
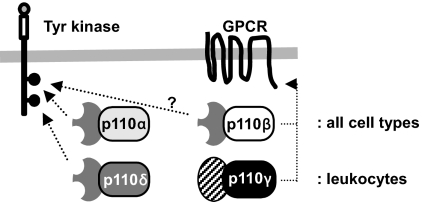
Taken together, our data suggest an analogy between p110α/p110δ and p110β/p110γ in the coverage of Tyr kinase- and GPCR-mediated PI3K signaling in distinct cell types (schematically shown in Fig. 4). Indeed, all evidence suggests that the ubiquitously expressed p110α plays an important role in Tyr kinase-driven PI3K signaling in all cell types (including leukocytes), with p110δ providing additional Tyr kinase-driven PI3K signaling in cell types in which it is expressed at high level, such as leukocytes. Under these conditions, the contribution of p110δ can also exceed that of p110α, for example in lymphocytes (ref. 32 and unpublished results). A similar scenario could be envisaged for the ubiquitously expressed p110β that could control GPCR-driven PI3K in all cell types, with p110γ providing additional GPCR-PI3K signaling capacity in white blood cells.
Fig. 4.
Model for class IA PI3K signaling downstream of Tyr kinase receptors and GPCRs. The ubiquitously expressed p110α plays an important role in Tyr kinase-driven PI3K signaling in all cell types, with p110δ providing additional Tyr kinase-driven PI3K signaling in cell types in which it is expressed at a high level, such as in leukocytes. A similar scenario could be envisaged for the broadly expressed p110β that could control GPCR-driven PI3K in all cell types, with p110γ providing additional GPCR-PI3K signaling capacity in white blood cells.
Materials and Methods
Reagents.
Small-molecule inhibitors were dissolved in DMSO, with final concentration of DMSO in the assays maximally 0.2%. LY294002 was from Calbiochem. The p110β inhibitor TGX-155 [in vitro IC50 for p110α: >20 μM; p110β: 0.03 μM; p110γ: >20 μM; p110δ: 0.34 μM (3, 33)] and p110γ inhibitor AS604850 [in vitro IC50 for p110α: 3.4 μM; p110β: >20 μM; p110γ: 0.19 μM; p110δ: >20 μM (11)] were a gift from Merck/Serono (Geneva). Abs to class IA PI3Ks were generated in-house. p110β Ab used for IP was generated by using a C-terminal peptide of p110β (KVNWMAHTVRKDYRS) as immunogen, affinity-purified over this peptide, and further purified over GST-p110α and GST-p110δ to deplete cross-reactivity with p110α and p110δ. p110β Abs used for immunoblot were from Santa Cruz Biotechnology (sc-602). These recognize the last 19 aa of p110β (SWTTKVNWMAHTVRKDYRS) and are referred to as p110β C-ter Abs. Anti-N-terminal p110β Abs used for Western blotting were generated in-house with p110β peptides present in exon 3, 5, 9, and 12 (QLEVPREATISY, EFRRKMRKFSEAKIQ, CKTKKSTKTINPSKYQ, and CLASGDSANVSSRGGK, respectively) as immunogen and used as a mix at 1 μg/ml each; this mix is referred as p110β Nter Abs. Additional PI3K Abs were from Upstate (p85pan; catalogue no. 06-195) or Alexis (p110γ; clone H1). Abs to Akt, α-tubulin, and GAPDH were from Cell Signaling, Sigma, and Abcam, respectively. Cell culture reagents were from Invitrogen. IGF1, EGF, SDF-1α, PDGF-BB, S1P, LPA, CSF-1, and C5a were from Preprotech. PTx (Bordetella pertussis) and 4-hydroxytamoxifen were from Sigma.
Mice.
Mice were kept in individually ventilated cages and cared for according to U.K. Home Office regulations. Constitutive conversion of the p110βflox gene into the p110βΔ21,22 allele was achieved by crossing p110βflox/flox mice with the Cre deleter strain. Cre-negative p110βΔ21,22/flox mice from a second generation were intercrossed to generate p110βΔ21,22/Δ21,22 and p110βflox/flox mice. Embryos from timed pregnant mice were used to prepare MEFs.
Fibroblast Culture and Stimulation.
Cells were cultured at 37°C in a humidified 5% CO2 atmosphere. MEFs were generated from embryonic day 13.5 embryos that were minced and dissociated with trypsin, and cells were allowed to adhere on dishes. NIH 3T3 cells and MEFs were cultured in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. For stimulation, NIH 3T3 cells and early-passage (P2–P5) MEFs were seeded in 10-cm dishes (106 cells per dish), starved in DMEM without FBS and antibiotics for 24 h, treated for 1 h with the indicated doses of TGX-155 (TG), AS604850 (AS), LY294002 (LY), or vehicle (DMSO), followed by stimulation with PDGF (30 ng/ml), insulin (1 μM), EGF (20 ng/ml), IGF1 (30 ng/ml), S1P (200 nM), SDF (30 ng/ml), or LPA (1 μM) for 10 min, unless stated otherwise. PTx (100 ng/ml) was added for 15 h in DMEM without FBS before cell stimulation.
Macrophage Culture and Stimulation.
Cells were cultured at 37°C in a humidified 5% CO2 atmosphere. BMMs were derived from bone marrow precursor cells isolated from femurs as detailed (34). PTx (active or heat-inactivated) was added 4 h before cell stimulation. PI3K inhibitors were included 30 min before cell stimulation. Cells were stimulated with CSF-1 (30 ng/ml) or C5a (100 ng/ml) for 5 min, unless stated otherwise.
Cell Transfection.
NIH 3T3 cells were transfected with pcDNA3 without or with the coding sequences for human p110γ (untagged) and porcine p101 (HA-tagged) by using Superfect (Qiagen), followed by selection in 800 μg/ml geneticin (Invitrogen) for at least 15 days. Geneticin-resistant clones from a single well were pooled and maintained under continuous selection in 200 μg/ml geneticin.
Immunoblotting.
Cell lysis and immunoblotting of NIH 3T3 cells and MEFs was performed as described (34). Total cell lysates of macrophages were prepared by directly harvesting cells in 2× Laemmli sample buffer, followed by shearing of DNA by passing five times through a 0.45-mm needle. Samples were resolved by 10% SDS/PAGE.
Lipid Kinase Assay.
PI3K activity assays were performed as described with PIP2 as a substrate.
Supplementary Material
Acknowledgments.
We thank A. Candi, P. Cutillas, P. Gonzalez-Gomez, A. Nitzsche, W. Pearce, E. Peskett, and C. See for help; S. Kulkarni and L. Stephens (Babraham Institute) for help in p110β mouse generation; E. Hirsch (University of Torino, Torino, Italy), M. Wymann (University of Basel, Basel, Switzerland), R. Wetzker (University of Jena, Jena, Germany), and C. Rommel (Merck-Serono) for p110γ reagents and KO mice; Merck-Serono and PIramed (Slough, U.K.) for small-molecule inhibitors; and members of the Center for Cell Signaling, especially M. Graupera, for feedback and support. This work was funded by Diabetes U.K. Grant BDA:RD 01/0002179, Biotechnology and Biological Science Research Council Grants BB/C505659/1 and BB/C50989/1, European Union Grant FP6-502935, and the Ludwig Institute for Cancer Research. J.G.-G. was supported by Fondation pour la Recherche Médicale Grant FRM-SPE20051105175, European Molecular Biology Organization Grant ALTF676–2005, and European Union Grant MEIF-CT-2006-039676). K.B. was supported by PIramed. F.R. was supported by a Medical Research Council capacity building award. K.O. was supported by Biotechnology and Biological Science Research Council David Phillips Fellowship JF1928.
Footnotes
Conflict of interest: B.V. is a consultant for PIramed (Slough, UK) and AstraZeneca.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707761105/DCSupplemental.
References
- 1.Vanhaesebroeck B, Leevers SJ, Panayotou G, Waterfield MD. Phosphoinositide 3-kinases: A conserved family of signal transducers. Trends Biochem Sci. 1997;22:267–272. doi: 10.1016/s0968-0004(97)01061-x. [DOI] [PubMed] [Google Scholar]
- 2.Bi L, Okabe I, Bernard DJ, Nussbaum RL. Early embryonic lethality in mice deficient in the p110β catalytic subunit of PI 3-kinase. Mamm Genome. 2002;13:169–172. doi: 10.1007/BF02684023. [DOI] [PubMed] [Google Scholar]
- 3.Jackson SP, et al. PI 3-kinase p110β: A new target for antithrombotic therapy. Nat Med. 2005;11:507–514. doi: 10.1038/nm1232. [DOI] [PubMed] [Google Scholar]
- 4.Kurosu H, et al. Heterodimeric phosphoinositide 3-kinase consisting of p85 and p110β is synergistically activated by the β-γ subunits of G proteins and phosphotyrosyl peptide. J Biol Chem. 1997;272:24252–24256. doi: 10.1074/jbc.272.39.24252. [DOI] [PubMed] [Google Scholar]
- 5.Maier U, Babich A, Nurnberg B. Roles of noncatalytic subunits in gbetagamma-induced activation of class I phosphoinositide 3-kinase isoforms β and γ. J Biol Chem. 1999;274:29311–29317. doi: 10.1074/jbc.274.41.29311. [DOI] [PubMed] [Google Scholar]
- 6.Graness A, Adomeit A, Heinze R, Wetzker R, Liebmann C. A novel mitogenic signaling pathway of bradykinin in the human colon carcinoma cell line SW-480 involves sequential activation of a Gq/11 protein, phosphatidylinositol 3-kinase β, and protein kinase C ε. J Biol Chem. 1998;273:32016–32022. doi: 10.1074/jbc.273.48.32016. [DOI] [PubMed] [Google Scholar]
- 7.Roche S, Downward J, Raynal P, Courtneidge SA. A function for phosphatidylinositol 3-kinase β (p85α-p110β) in fibroblasts during mitogenesis: Requirement for insulin- and lysophosphatidic acid-mediated signal transduction. Mol Cell Biol. 1998;18:7119–7129. doi: 10.1128/mcb.18.12.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kubo H, Hazeki K, Takasuga S, Hazeki O. Specific role for p85/p110β in GTP binding protein-mediated activation of Akt. Biochem J. 2005;392:607–614. doi: 10.1042/BJ20050671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yart A, et al. A function for phosphoinositide 3-kinase β lipid products in coupling β γ to Ras activation in response to lysophosphatidic acid. J Biol Chem. 2002;277:21167–21178. doi: 10.1074/jbc.M110411200. [DOI] [PubMed] [Google Scholar]
- 10.Brachmann SM, Ueki K, Engelman JA, Kahn RC, Cantley LC. Phosphoinositide 3-kinase catalytic subunit deletion and regulatory subunit deletion have opposite effects on insulin sensitivity in mice. Mol Cell Biol. 2005;25:1596–1607. doi: 10.1128/MCB.25.5.1596-1607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camps M, et al. Blockade of PI3Kγ suppresses joint inflammation and damage in mouse models of rheumatoid arthritis. Nat Med. 2005;11:936–943. doi: 10.1038/nm1284. [DOI] [PubMed] [Google Scholar]
- 12.Hirsch E, et al. Central role for G protein-coupled phosphoinositide 3-kinase γ in inflammation. Science. 2000;287:1049–1053. doi: 10.1126/science.287.5455.1049. [DOI] [PubMed] [Google Scholar]
- 13.Jones GE, et al. Requirement for PI 3-kinase γ in macrophage migration to MCP-1 and CSF-1. Exp Cell Res. 2003;290:120–131. doi: 10.1016/s0014-4827(03)00318-5. [DOI] [PubMed] [Google Scholar]
- 14.Vanhaesebroeck B, et al. Distinct PI(3)Ks mediate mitogenic signaling and cell migration in macrophages. Nat Cell Biol. 1999;1:69–71. doi: 10.1038/9045. [DOI] [PubMed] [Google Scholar]
- 15.Geering B, Cutillas PR, Nock G, Gharbi SI, Vanhaesebroeck B. Class IA phosphoinositide 3-kinases are obligate p85–p110 heterodimers. Proc Natl Acad Sci USA. 2007;104:7809–7814. doi: 10.1073/pnas.0700373104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patrucco E, et al. PI3Kγ modulates the cardiac response to chronic pressure overload by distinct kinase-dependent and -independent effects. Cell. 2004;118:375–387. doi: 10.1016/j.cell.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 17.Seibler J, et al. Rapid generation of inducible mouse mutants. Nucleic Acids Res. 2003;31:e12. doi: 10.1093/nar/gng012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasaki T, et al. Function of PI3Kγ in thymocyte development, T cell activation, and neutrophil migration. Science. 2000;287:1040–1046. doi: 10.1126/science.287.5455.1040. [DOI] [PubMed] [Google Scholar]
- 19.Laffargue M, et al. Phosphoinositide 3-kinase γ is an essential amplifier of mast cell function. Immunity. 2002;16:441–451. doi: 10.1016/s1074-7613(02)00282-0. [DOI] [PubMed] [Google Scholar]
- 20.Foukas LC, et al. Critical role for the p110α phosphoinositide-3-OH kinase in growth and metabolic regulation. Nature. 2006;441:366–370. doi: 10.1038/nature04694. [DOI] [PubMed] [Google Scholar]
- 21.Knight ZA, et al. A pharmacological map of the PI3-K family defines a role for p110α in insulin signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hooshmand-Rad R, et al. The PI 3-kinase isoforms p110(α) and p110(β) have differential roles in PDGF- and insulin-mediated signaling. J Cell Sci. 2000;113:207–214. doi: 10.1242/jcs.113.2.207. [DOI] [PubMed] [Google Scholar]
- 23.Park CS, Schneider IC, Haugh JM. Kinetic analysis of platelet-derived growth factor receptor/phosphoinositide 3-kinase/Akt signaling in fibroblasts. J Biol Chem. 2003;278:37064–37072. doi: 10.1074/jbc.M304968200. [DOI] [PubMed] [Google Scholar]
- 24.Christoforidis S, et al. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol. 1999;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- 25.Meier TI, et al. Cloning, expression, purification, and characterization of the human Class Ia phosphoinositide 3-kinase isoforms. Protein Expression Purif. 2004;35:218–224. doi: 10.1016/j.pep.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 26.Tang X, Downes CP. Purification and characterization of Gbetagamma-responsive phosphoinositide 3-kinases from pig platelet cytosol. J Biol Chem. 1997;272:14193–14199. doi: 10.1074/jbc.272.22.14193. [DOI] [PubMed] [Google Scholar]
- 27.Luttrell DK, Luttrell LM. Not so strange bedfellows: G protein-coupled receptors and Src family kinases. Oncogene. 2004;23:7969–7978. doi: 10.1038/sj.onc.1208162. [DOI] [PubMed] [Google Scholar]
- 28.Leverrier Y, et al. Class I phosphoinositide 3-kinase p110β is required for apoptotic cell and Fcγ receptor-mediated phagocytosis by macrophages. J Biol Chem. 2003;278:38437–38442. doi: 10.1074/jbc.M306649200. [DOI] [PubMed] [Google Scholar]
- 29.Hill K, et al. Specific requirement for the p85–p110α phosphatidylinositol 3-kinase during epidermal growth factor-stimulated actin nucleation in breast cancer cells. J Biol Chem. 2000;275:3741–3744. doi: 10.1074/jbc.275.6.3741. [DOI] [PubMed] [Google Scholar]
- 30.Smith AJ, et al. p110β and p110δ phosphatidylinositol 3-kinases up-regulate Fc(ε)RI-activated Ca2+ influx by enhancing inositol 1,4,5-trisphosphate production. J Biol Chem. 2001;276:17213–17220. doi: 10.1074/jbc.M100417200. [DOI] [PubMed] [Google Scholar]
- 31.Chaussade C, et al. Evidence for functional redundancy of class-IA PI 3-kinase isoforms in insulin signaling. Biochem J. 2007;404:449–458. doi: 10.1042/BJ20070003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okkenhaug K, et al. Impaired B and T cell antigen receptor signaling in p110δ PI 3-kinase mutant mice. Science. 2002;297:1031–1034. doi: 10.1126/science.1073560. [DOI] [PubMed] [Google Scholar]
- 33.Ali K, et al. Isoform-specific functions of phosphoinositide 3-kinases: p110δ but not p110γ promotes optimal allergic responses in vivo. J Immunol. 2007;180:2538–2544. doi: 10.4049/jimmunol.180.4.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papakonstanti EA, Ridley AJ, Vanhaesebroeck B. The p110δ isoform of PI 3-kinase negatively controls RhoA and PTEN. EMBO J. 2007;26:3050–3061. doi: 10.1038/sj.emboj.7601763. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.