Abstract
Cohesin is required to prevent premature dissociation of sister chromatids after DNA replication. Although its role in chromatid cohesion is well established, the functional significance of cohesin's association with interphase chromatin is not clear. Using a quantitative proteomics approach, we show that the STAG1 (Scc3/SA1) subunit of cohesin interacts with the CCTC-binding factor CTCF bound to the c-myc insulator element. Both allele-specific binding of CTCF and Scc3/SA1 at the imprinted IGF2/H19 gene locus and our analyses of human DM1 alleles containing base substitutions at CTCF-binding motifs indicate that cohesin recruitment to chromosomal sites depends on the presence of CTCF. A large-scale genomic survey using ChIP-Chip demonstrates that Scc3/SA1 binding strongly correlates with the CTCF-binding site distribution in chromosomal arms. However, some chromosomal sites interact exclusively with CTCF, whereas others interact with Scc3/SA1 only. Furthermore, immunofluorescence microscopy and ChIP-Chip experiments demonstrate that CTCF associates with both centromeres and chromosomal arms during metaphase. These results link cohesin to gene regulatory functions and suggest an essential role for CTCF during sister chromatid cohesion. These results have implications for the functional role of cohesin subunits in the pathogenesis of Cornelia de Lange syndrome and Roberts syndromes.
Keywords: cohesion, transcription, insulator, centromere, metaphase
In eukaryotes, gene loci are often clustered together to form nuclear territories characterized by specific expression profiles (1, 2). Insulator and boundary elements are thought to contribute to the functional identity of genes and territories by shielding individual genomic regions from the influence of neighboring enhancer or silencer elements (3). Although the exact mechanism by which insulators achieve a selective interaction of enhancers with specific promoters is still controversial, evidence from studies in Drosophila and vertebrates suggests that the formation of chromatin loops may be the underlying principle of insulator function (3, 4). For instance, multiple gypsy insulator elements form “insulator bodies” that attach to the nuclear membrane and fold chromosomes into loops (4). In vertebrates, the highly conserved CCTC-binding factor CTCF is the only known factor with insulator function. CTCF mediates long-range chromatin interactions between loci at the β-globin gene in erythroid cells and at the IGF2/H19 gene loci (5). Evidence suggests that the ability of CTCF to interact with itself contributes to the formation of “active chromatin hubs” at these loci (6).
CTCF has been assigned a wide variety of functions, and their diversity reflects the number of potential interacting partners (3, 7, 8). Although the association of CTCF with YB1, YY1, Kaiso, and Sin3 may confer transcription regulator activity, interaction of CTCF with both nucleophosmin and/or chromodomain helicase CHD8 is required for insulator function (6, 9). However, it remains unclear whether these interactions are required at all genomic CTCF sites or whether they regulate a subset of insulator elements.
Analyses of the genome-wide distribution of CTCF revealed a tendency of CTCF to reside in loci that separate presumptive chromosomal domains (10, 11), consistent with the requirement to preserve functional integrity by limiting the spread of regulatory signals emanating from neighboring domains. CTCF is present at the boundary between genes that are subject to X inactivation and genes that escape this silencing (11, 12). CTCF is also an integral component of the c-myc insulator element MINE that separates the transcriptionally active c-myc gene from surrounding chromatin that bears features typical for heterochromatin (13). The mode of action of CTCF at this genomic region, however, is unclear. Here, we report the identification of Scc3/SA1, a subunit of the cohesin complex, as an interacting partner for CTCF. Our investigations both identify the previously unknown mechanism of chromatin–cohesin interaction and suggest a role for CTCF in the coordinated distribution of replicated DNA during mitosis.
Results
Detection of CTCF Interacting Proteins by Isotope Coded Affinity Tag (ICAT) Quantitative Mass Spectrometry.
To define the composition of the CTCF complex that mediates insulator activity at the mammalian c-myc gene, we applied a quantitative proteomics approach (14). This strategy is based on the use of stable isotope tagging and mass spectrometry to distinguish proteins that are specifically enriched in a purification procedure from copurifying contaminants [supporting information (SI) Fig. S1]. For this analysis, we compared the enrichment ratios of nuclear extract proteins that associate with immobilized templates containing either the wild-type or a mutated CTCF-binding site at the c-myc insulator element (Fig. S1). Initial in vitro affinity purifications and subsequent Western analysis demonstrated that the DNA template with the wild-type CTCF sequence recruits high levels of CTCF, whereas three point mutations within the target sequence resulted in a significant reduction in CTCF recruitment (Fig. S1). To identify cofactors recruited by CTCF, we differentially labeled components bound to the wild-type and mutant sequences with heavy and light ICAT reagents (14). The labeled protein pools were combined and proteolyzed. After fractionation and identification of the peptides by mass spectrometry, we calculated the relative abundance of heavy- and light-labeled peptides/proteins. Applying this experimental approach, we identified 9,605 peptides (P > 0.99), corresponding to 2,084 proteins (P > 0.9). CTCF-derived peptides were identified by 112 independent spectra and were 3- to 25-fold enriched in the protein fraction purified using the wild-type CTCF-binding site (average 7.7-fold, P = 3.1e-03). The vast majority of peptides identified bound nonspecifically, as is evident by their lack of enrichment in either protein pool (Fig. S2). However, applying an enrichment ratio threshold of at least 2-fold and a P value threshold <0.01 revealed that the nuclear protein Scc3/SA1 (STAG1) preferentially associated with DNA containing the CTCF target sequence of the c-myc insulator element (P = 5.8e-03; Fig. 1). Scc3/SA1 and the much more abundant isoform Scc3/SA2 (STAG2) participate in the formation of the vertebrate cohesin complex in a mutually exclusive manner (15, 16).
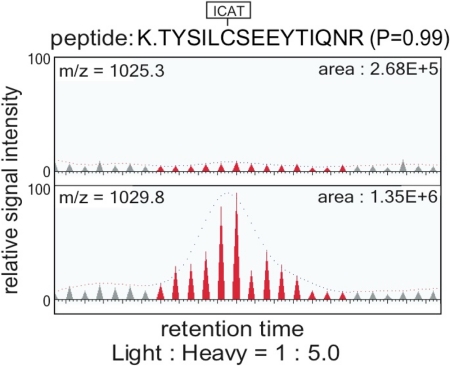
Fig. 1.
Detection of CTCF-interacting factors by ICAT quantitative mass spectrometry. Protein samples recovered from wild-type and mutant CTCF-binding sites (Fig. S1) were labeled with the heavy and normal ICAT reagents, respectively, and prepared for μLC-MS/MS (14). SEQUEST database searching matched an MS/MS spectrum to the indicated ICAT-labeled peptide sequence derived from Scc3/SA1 as the best match. The relative abundance of isotopically heavy (Lower) and normal (Upper) ICAT-labeled peptides corresponding to Scc3/SA1 was calculated by reconstructing single-ion chromatograms for each peptide using XPRESS (14). A peptide derived from the Scc3/SA1 was enriched to a similar level (1:5.0) as peptides corresponding to CTCF (1:7.7).
To confirm the quantitative enrichment of Scc3/SA1 in DNA-bound CTCF complexes, we performed Western blot analysis experiments on proteins purified from nuclear extracts using the immobilized templates. Templates containing the CTCF wild-type target sequence recovered ≈3-fold higher levels of Scc3/SA1 (data not shown), supporting the results obtained by mass spectrometry.
In Vivo Colocalization and Genomic Distribution of Scc3/SA1 and CTCF.
Our quantitative mass spectrometry approach suggests that both CTCF and the cohesin subunit Scc3/SA1 are recruited to the c-myc insulator upstream sequence. To test whether this correlation also holds true in vivo, we performed ChIP experiments with a human T cell line (Jurkat). We tested 11 regions across the human c-myc gene domain, four of which are known to associate with CTCF (refs. 10 and 11 and our data not shown). All four regions that associate with CTCF also associate with Scc3/SA1 (Fig. S3). To determine the genome-wide significance of our observation, we conducted a more comprehensive analysis of Scc3/SA1 binding in HBL100 cells across the nonrepetitive sequences of ENCODE regions representing 1% of the human genome. DNA recovered from the anti-CTCF and -Scc3/SA1 immunoprecipitates was amplified, labeled, and hybridized to tiling arrays. ChIP-Chip was performed in triplicate using independently prepared biological samples, as described in Methods. To determine the degree of overlap, the genomic positions and intensities of putative binding sites were identified for both CTCF and Scc3/SA1 at a high resolution using MeDiChI, a regression-based procedure that learns a generative model of joint (multiple, potentially overlapping) binding events in normalized ChIP-Chip data (17). Using a binding-site P value cutoff of 0.01, MeDiChI identified 147 CTCF sites (Fig. 2A and Table S1). A large percentage (86%, 126 of 147) overlaps with sites previously identified in a genome-wide analysis of CTCF binding in IMR90 cells (11). Furthermore, a search for motifs in these 147 sites using MEME (18) revealed that 63% of sequences contain a motif highly similar to the CTCF motif reported (Fig. S4; ref. 11). The high level of concordance between CTCF-binding site distributions in both HBL100 and IMR90 cells and the presence of the CTCF motif in a similar percentage of binding sequences demonstrate that our ChIP-Chip approach identified CTCF sites with high accuracy.
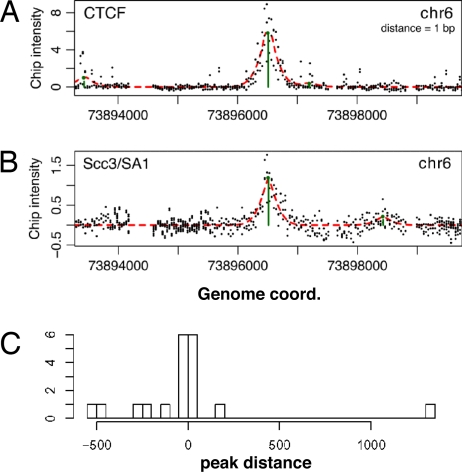
Fig. 2.
Detection and colocalization of CTCF and Scc3/SA1 in ChIP-Chip experiments. (A) Plot of intensity ratios of individual probes within a region of chromosome 6 and detection of a CTCF-binding site by MeDiCHi (see Methods). (B) Plot of intensity ratios of the Scc3/SA1 ChIP-Chip experiment within the same chromosomal region as in A. (C) Frequency histogram of the distances of Scc3/SA1 peaks (P ≤ 0.01) to all CTCF peaks (P ≤ 0.01) within 2,000 bp.
Applying the same experimental approach using the anti-Scc3/SA1 antibody, we identified 41 Scc3/SA1-binding sites within the ENCODE genomic region (P = 0.01; Fig. 2B and Table S2). To define the degree of CTCF-Scc3/SA1 colocalization, the minimum distances of the 147 CTCF-binding sites to any of the Scc3/SA1-binding sites were computed (Fig. 2C). We found that 16 of the 41 Scc3/SA1-binding sites occur within 1,000 bp of at least one CTCF site (Table S3). Simulations of chosen pairs of hits in the ENCODE region showed that this number of small distances has a negligible probability of occurring by chance (P < 10−8). Thus, colocalization of Scc3/SA1 and CTCF at a subset of CTCF binding sites is highly significant and not the result of a random event.
Allele-Specific Binding of Scc3/SA1 at the Imprinted IGF2/H19 Gene Region.
CTCF participates in the allele-specific regulation of gene expression at the imprinted IGF2/H19 gene locus. The methylation of cytosine residues and concomitant recruitment of tri-methylated histone H3 at the imprinting control region (ICR) of the paternal allele inhibits the association of CTCF, resulting in transcriptional enhancement of the paternal IGF2 gene through enhancers located near the H19 gene. In contrast, the lack of ICR methylation at the maternal allele allows CTCF to bind multiple sites upstream of the H19 gene, subsequently blocking the interaction of the enhancers and the IGF2 promoter. To determine whether Scc3/SA1 also binds to the CTCF sites at the IGF2/H19 gene locus in an allele-specific manner, we determined the allele-specific association of Scc3/SA1, CTCF and lysine 9-trimethylated histone H3 (H3K9me3) using methylation-sensitive PCR. Chromatin from HBL100 cells was immunoprecipitated with antibodies specific for Scc3/SA1, CTCF, and H3K9me3 (Fig. 3). DNA from the immunoprecipitates was split into two aliquots and digested with either the methylation-sensitive restriction enzyme AciI or with EcoRI that is unaffected by CpG methylation. Subsequent PCR amplification with primers surrounding the ICR distinguished between methylated and nonmethylated IGF2 alleles. DNA recovered from ChIP with an H3K9me3 antibody remained unaffected by AciI digestion (Aci I signal/EcoRI signal = 97%), confirming that this histone modification is associated with the methylated cytosine on the paternal allele. In contrast, DNA recovered from ChIP with a CTCF antibody was highly sensitive to AciI, as indicated by the significant loss of amplified DNA after treatment with AciI. Similarly, PCR amplification of AciI-digested DNA recovered from anti-Scc3/SA1 immunoprecipitates resulted in only 14% of signal relative to EcoRI-digested DNA (Fig. 3). Thus, in concordance with a previous report (19), our data demonstrate that both CTCF and Scc3/SA1 bind in an allele-specific manner to the nonmethylated ICR on the maternal allele.
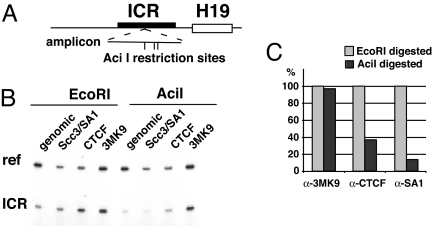
Fig. 3.
Allele-specific association of cohesin and CTCF with the Igf2/H19 ICR is detected by methylation-sensitive PCR. (A) Position of AciI restriction sites within the amplified region of the ICR. (B) The CpG methylation sensitive restriction enzyme AciI, but not EcoRI, reduces the number of genomic templates available for PCR amplification of the ICR recovered after chromatin immunoprecipitation with either anti-CTCF or -Scc3/SA1 but not after immunoprecipitation with anti-3mK9 H3 (histone H3 trimethylated at lysine 9). As an internal control, a genomic region of the beta-globin gene promoter that lacks both EcoRI and AciI sites was coamplified. (C) Quantitation of the methylation sensitive PCR. Ratios of ICR/beta-globin signals were determined for each immunoprecipitation (dark bars) and compared with the signal ratio after digestion with EcoRI, which was set to 100. Although the paternal allele is CpG methylated and associated with histone H3 trimethylated at lysine 9, the maternal allele is unmethylated and associated with CTCF and Scc3/SA1.
CTCF Is Required for Scc3/SA1 Recruitment to Chromatin.
Our comparative ChIP-Chip analysis revealed a nonrandom distribution of CTCF and Scc3/SA1 binding across the human genome. To test whether the association of Scc3/SA1 with chromatin directly depended on the presence of CTCF, we analyzed Scc3/SA1 binding at both a normal and a mutated CTCF-dependent insulator element of the human myotonic dystrophy gene DM1 that had been integrated at the same genomic locus in mouse 3T3 cells via recombination-mediated cassette exchange (RMCE; ref. 20). The DM1 alleles under study are identical except in sequences at the CTCF-binding sites that contain base substitutions to eliminate CTCF binding (ref. 20; Fig. S5). ChIP analysis of the normal allele of the insulator fragment demonstrated that both CTCF and Scc3/SA1 were bound at the CTCF-binding site of the normal allele of the insulator element (Fig. 4). In contrast, base mutations introduced into the CTCF target sequence of the DM1 insulator abolished CTCF binding, as observed by the loss of enrichment after ChIP with anti-CTCF antibodies. Importantly, point mutations of CTCF-binding sequences also abrogated the association of Scc3/SA1 with the human DM1 allele. These data demonstrate that cohesin binding at the DM1 gene directly depends on the presence of CTCF.
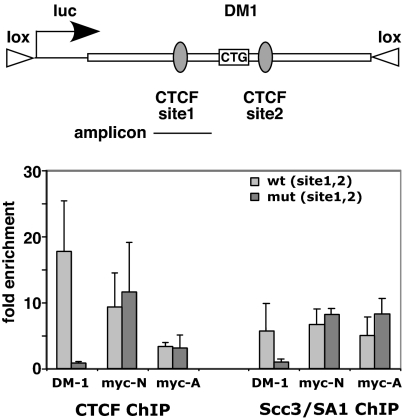
Fig. 4.
Association of cohesin with the myotonic dystrophy gene DM1 requires CTCF. (A) Scheme of the human DM1 sequences integrated via RMCE into murine 3T3 cells. Sequences of the wild-type and mutant CTCF-binding sites 1 and 2 are shown in Fig. S5. (B) ChIP experiments at the wild-type and mutant DM1 loci reveal that mutations in CTCF motifs 1 and 2 [mut (site1,2)] abrogate binding of both CTCF and Scc3/SA1, whereas binding of CTCF and Scc3/SA1 to myc-N and myc-A, both binding sites for CTCF at the endogenous murine c-myc gene (8, 13), remains unaffected. Bars represent the average and standard deviation of three independent experiments.
CTCF-Dependent and Independent Association of Scc3/SA1 with Chromosomal Sites.
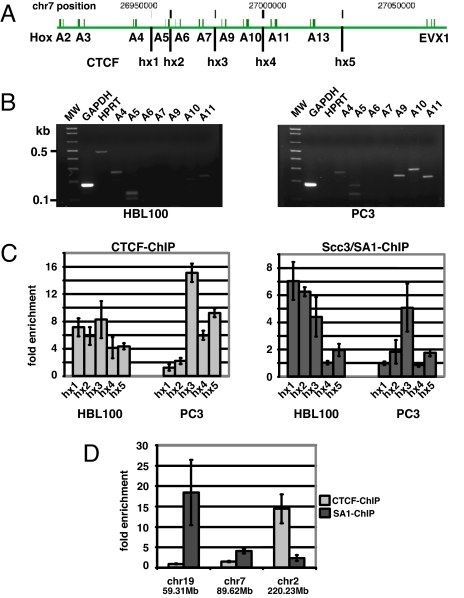
Our survey of the Scc3/SA1-binding site distribution within the ENCODE sequences revealed a significant coincidence of CTCF and Scc3/SA1 binding, and this colocalization depends on the presence of CTCF. However, only 16 of the 147 CTCF-binding sites within ENCODE were also bound by Scc3/SA1. To determine whether the lower number of Scc3/SA1 sites relative to CTCF sites was due to selective binding of Scc3/SA1 or simply to experimental variables (e.g., lower efficiency of the anti-Scc3/SA1 antibody in immunoprecipitations), we chose to study the HoxA gene domain by conventional ChIP techniques. The HoxA domain harbors multiple genes, which are expressed according to the rules of colinearity, with 3′ Hox genes (e.g., hoxA1 to hoxA4) expressed early in development and 5′ HoxA genes (e.g., HoxA10 to HoxA13) later in development (21). Accordingly, 3′ HoxA genes are preferentially expressed in the HBL100 breast epithelial cell line, whereas 5′ HoxA genes are preferentially expressed in the PC3 prostate cell line. In our ChIP-Chip analysis, we identified five genomic regions within the HoxA locus in HBL100 cells that bind CTCF (hx1–5, Fig. 5). Using quantitative PCR analyses, we largely confirmed binding of CTCF at hx1–5 in the HoxA locus of HBL100 and PC3 cells, except that CTCF-binding sites at hx1 and hx2 were not significantly enriched in PC3 cells. Importantly, whereas hx4 associated with CTCF in both HBL100 and PC3 cells, it failed to recruit Scc3/SA1. These results demonstrate that Scc3/SA1 selectively associates with a subset of CTCF-bound regions.
Fig. 5.
Selective association of Scc3/SA1 with genomic regions. (A) Schematic description of the human HoxA locus. CTCF-binding sites are indicated by vertical bars hx1–hx5. (B) Expression pattern of Hox genes A2–A11 in HBL100 and PC3 cells. Expression of GAPDH is used as a reference. (C) Relative enrichment of genomic regions hx1–hx5 after ChIP with anti-CTCF or -Scc3/SA1. Site hx4 associates with CTCF but does not recruit Scc3/SA1. Relative enrichments are representative data from three independent experiments. (D) ChIP reveals selective binding of CTCF and Scc3/SA1 to genomic regions on chromosome 7 [position 89620788, hg17, University of California, Santa Cruz (UCSC)], chromosome 19 (position 59310952, hg17, UCSC), and chromosome 2 (position 220232167, hg17, UCSC). Genomic regions on chromosomes 7 and 19 associate with Scc3/SA1 but not with CTCF. In contrast, position 220232167 on chromosome 2 binds CTCF but not Scc3/SA1. Error bars indicate standard deviation of enrichment observed in three independent ChIP.
In addition to genomic sites that exclusively bind CTCF, the large-scale survey of Scc3/SA1-associated sites within ENCODE sequences also revealed 25 positions that failed to correspond to CTCF-bound sequences within 1 kb of flanking sequences on either side. Of these, we selected 11 sites for testing with conventional ChIP-PCR. Although the majority of these regions were false positives, two sites within the ENCODE region on chromosome 19 (chr19, 59310952, Table S2) and chromosome 7 (chr7, 89620788, Table S2) were indeed bound by Scc3/SA1 but not by CTCF (Fig. 5D). These data suggest that a subset of chromosomal sites use alternative mechanisms to recruit cohesin to chromatin.
CTCF Associates with Centromeric Regions.
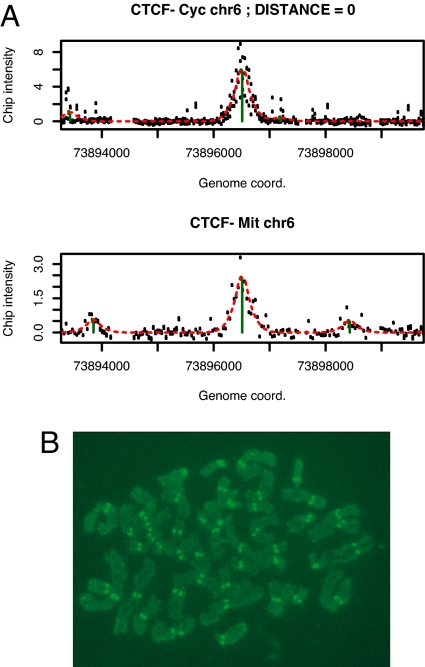
In mitotic metaphase, cohesin is essential for sister chromatid cohesion (15). Although the bulk of cohesin is removed from chromosome arms during prophase and prometaphase, the sister chromatids remain attached through centromeres (22, 23). To address the question of whether CTCF remains associated with cohesin during mitosis, we used ChIP-Chip on mitotic chromatin preparations (Fig. S6). Our results confirm previous reports that a large fraction of CTCF remains associated with chromosomes during metaphase (24). Seventy of 147 CTCF sites detected in asynchronously growing HBL100 cells were also found in chromatin of mitotic HBL100 cells (Fig. 6A and Table S4). Because centromeric DNA consists of large arrays of repetitive DNA, our ChIP-Chip approach was technically unsuited for detection of an association of CTCF with centromeres during metaphase. Therefore, we performed immunofluorescence detection of CTCF in metaphase chromosomes. Paraformaldehyde-cross-linked metaphase chromosome preparations were incubated with anti-CTCF and detected by FITC-linked secondary anti-rabbit antibodies. Consistent with previous observations of CTCF distribution during mitosis (24), we found a strong and punctuate fluorescence label at the centromeric regions of metaphase chromosomes (Fig. 6). Although these observations suggest that the presence of CTCF is required for cohesin function in later stages of mitosis, additional experiments will be required to define the precise role for CTCF in linking cohesin to chromatin both during interphase and the mitotic phase of the cell cycle.
Fig. 6.
Association of CTCF with chromosomal sites and centromeres during mitosis. (A) Example of a CTCF site on chromosome 6 occupied in both asynchronously growing cells (Upper, CTCF Cyc) and cells arrested in mitosis (Lower, CTCF-Mit). The calculated distance between the peaks is indicated (distance = 0). A complete list of all sites detected in both asynchronously growing and mitotic cells is shown in Table S4. (B) Immunofluorescence detection of CTCF bound to centromeres of human mitotic chromosomes. A representative example of a mitotic chromosome preparation is shown. All cells examined from each preparation showed centromeric staining by CTCF.
Discussion
Our discovery of the interaction of CTCF with cohesin offers previously unsuspected explanations for the regulatory role of enhancer-promoter communications in vertebrates. Although the requirement of cohesin for sister chromatid cohesion after DNA replication during S-phase is well documented, its function in transcription regulation is unknown (25). Previous studies in yeast, Drosophila, and zebrafish suggest that cohesin influences transcription. For example, in Saccharomyces cerevisiae, cohesin supports the establishment and maintenance of boundaries at the HMR-silent mating type loci. Mutations in the SMC1 component of cohesin cause a loss of HMR boundary activity, with resultant silencing of adjacent genomic regions (26). In addition, genetic screens in Drosophila have identified Nipped-B/Scc2, an ortholog of the yeast cohesin loading complex Scc2/Scc4, as an important factor in enhancer-promoter communication at the cut gene (27). The effect of mutated Nipped-B was found to be most severe when a gypsy insulator element was present between the enhancer and the promoter. Subsequent RNAi knockdown experiments to reduce the cellular levels of Nipped-B or Scc3/SA1 suggest that these two members of the cohesin complex have opposing effects on cut gene activity (28). Although reduced Nipped-B levels decrease cut gene activity, reduced Scc3/SA1 levels increase cut transcription. Similarly, cohesin-dependent gene regulation has been shown for the runx genes in zebrafish (29). A genetic screen revealed that embryos lacking Rad21, the Scc1 subunit of cohesin, fail to develop differentiated blood cells because of a lack of runx3 expression.
A clinically relevant example of a role of cohesin in regulating transcription and/or enhancer activities comes from the observation of mutations in subunits of the cohesin complex that can severely affect normal human development. Cornelia de Lange (CdLS) syndrome, characterized by developmental anomalies including limb anomalies, short stature, and mental retardation, is most frequently caused by mutations in the human ortholog of Nipped-B, the nipped-B-like gene NIPBL, or delangin (30, 31). Some affected individuals have been found to carry mutant forms of SMC1 or SMC3, both integral members of the cohesin complex (32, 33). Although the relative contribution of defects in transcription as opposed to defects of sister chromatid cohesion in CdLS remains unknown, our data, together with the known role of CTCF in enhancer insulation, support the idea that the severe phenotype seen in CdLS results from a defect in the function of cohesin and CTCF in regulating gene activity.
The ability of cohesin to “glue” together two DNA strands clearly lends itself to the idea that the highly organized arrangement of chromosomal territories is at least partly guided by CTCF and cohesin. CTCF has previously been shown to participate in intra- and interchromosomal looping at the β-globin and Igf2/H19 gene loci (34–36). The ability of CTCF to both form dimers in vitro and interact with itself in yeast two-hybrid experiments is consistent with its role in promoting long-range chromatin interactions (3, 6). Our finding that CTCF interacts with cohesin provides an additional or alternative mechanism that may be the underlying principle of CTCF-mediated long-range interaction of genomic regions. The cohesin subunits SMC1 and SMC3 associate with Scc1/Rad21 and Scc3/SA to form a ring-like structure that is capable of holding together two DNA strands replicated during S-phase (37). Although the exact structural and functional roles of cohesin binding to chromosomes during interphase still need to be addressed, our results strongly suggest that the interaction of cohesin with a subset of CTCF-associated regions is involved in the generation of intra- and interchromosomal association of genomic regions.
Cohesion of sister chromatids during mitosis requires the cohesin complex. Previous observations have highlighted differences among cohesin complexes associated with centromeric regions or chromosome arms (23, 38). Although the bulk of cohesin complexes dissociates from chromosomal arms during prophase and prometaphase, a fraction remains associated preferentially at centromeric regions of chromosomes, and mediates cohesion of replicated DNA strands promoting the equal distribution of sister chromatids to daughter cells in mitosis. The mechanisms of both recruitment and dissociation of cohesin during later stages in mitosis still remain relatively obscure. The results of this study suggest that cohesin is recruited to chromosomal arms and to centromeres, possibly via its interaction with CTCF. Thus, future models that address the functional mechanism of cohesin dissociation during mitosis will have to consider the potential of CTCF posttranslational modifications for the release of sister chromatid cohesion.
While this manuscript was in preparation, two other reports were published confirming our observation that cohesin both colocalizes with and requires CTCF for binding to chromatin (39, 40). Our studies confirm and extend those observations by demonstrating that base substitutions within the CTCF-motif at the DM1 locus abolish both CTCF and cohesin binding. Furthermore, our data suggest that this interaction likely occurs via Scc3/SA1, because only Scc3/SA1 but not Rad21 or SMC1/SMC3 showed significant enrichment in our proteomic analysis of proteins bound to immobilized templates. Finally, our immunofluorescence data demonstrate that CTCF associates with centromeric heterochromatin during mitosis, consistent with the high concordance of CTCF distribution in cycling and mitotically arrested cells (Fig. 6A, Table S4, and ref. 24). Thus, in combination, our data suggest that CTCF is required for cohesin binding in interphase and mitotic cells.
Methods
Protein Purification Using Immobilized DNA Templates, ICAT Labeling, and Quantitative Mass Spectrometry.
Crude nuclear extract was prepared from 109 Jurkat cells grown in growth media (RPMI medium 1640/10% FBS) according to ref. 41. Template DNA was generated by PCR amplification of a 163-bp region derived from a normal and mutant c-myc insulator sequences (Fig. S1) using a biotinylated/nonbiotinylated primer combination. To generate the immobilized templates, 175 pmols of either wild-type or mutant PCR-amplified DNA was coupled with 8.75 mg of streptavidin-linked magnetic beads (Dynal M280) essentially as described (42).
Forty milligrams of nuclear extract prepared from Jurkat cells was incubated with immobilized templates for 2 h at 4°C. To reduce nonspecific binding of nuclear proteins to nucleic acid, each binding reaction contained 350 pmols of nonbiotinylated 163-bp mutant DNA (CTCF-mut). After four washes of the template-bound complexes, proteins were eluted with elution buffer (5 mM Tris·HCl, pH 7.5, 0.5 mM EDTA, 1 M NaHCO3). Labeling of eluted proteins with ICAT reagents was performed as described (42, 43).
μLC-MS/MS and Data Analysis.
μLC-MS/MS was essentially done as described (44). Cysteine-containing tryptic peptides were identified by searching MS/MS spectra against a human protein sequence database using SEQUEST as described (45). Data were quantified and analyzed essentially as described by using the XPRESS and INTERACT computer programs, respectively (43). Data were filtered by using a requirement for peptides tryptic at both N and C termini. PeptideProphet and ProteinProphet (14) were used to determine the probability that peptide and protein assignments were correct. A probability cutoff value of 0.99 and 0.9 was used for the respective analysis.
ChIP.
Chromatin was prepared for immunoprecipitation as described (13) by cross-linking the cells in 1% formaldehyde for 5 min and subsequent sonication until the bulk of DNA was 300–600 bp in size. Chromatin corresponding to 2 × 107 cells was immunoprecipitated with anti-CTCF antibody (Upstate Biotechnology, 06-917), anti-Scc3/SA1 antibody (Abcam, #4457), or anti-trimethyl-Histone-H3 (3MK9, Upstate Biotechnology, #07-442). Immunoprecipitates were washed, the DNA-protein cross-links reversed, and the recovered DNA was tested in regular conventional quantitative PCR as described (13). Sequences of primers specific for gene loci under study and reference primers (β-globin) are available upon request.
ChIP-Chip Analyses.
The amplification and preparation of immunoprecipitated DNA for hybridizations to ENCODE arrays (NimbleGen Systems) were performed essentially as described in ref. 46 with minor modifications at DNA preparation and purification steps. Briefly, immunoprecipitated DNA was end-repaired by using T4 DNA polymerase at 12°C for 15 min (New England Biolabs, M0203S) according to the manufacturer's recommendation. After phenol/chloroform purification and ethanol precipitation, the end-repaired DNA was ligated to linker-primer with commercially available DNA ligation kits (Quick Ligation kit M2200S, New England Biolabs). DNA was amplified with twenty PCR cycles and purified with centrifugal filter units (Microcon YM-50) according to the manufacturer's protocol. PCR amplicons were further amplified by subsequent PCRs using 50 ng of DNA to obtain a sufficient amount of DNA required for DNA array hybridization.
Sample labeling, array hybridization, and determination of relative probe intensities was performed at NimbleGen.
Peak Detection and Analysis.
Binding sites in both the CTCF and Scc3/SA1 data were detected by using the MeDiChI (17) statistical software, which learns a model of the binding peak profile for each dataset, and then fits that profile to the reference-normalized (Cy5/Cy3) probe intensities using a constrained linear model. The resulting coefficients of the fitted model contain the high-resolution best-fit genomic coordinates and intensities of each detectable peak in the dataset (within the ENCODE region). The P value for a given detection was estimated from the result of 1,000 bootstrap runs of the same procedure on the data residuals (i.e., probe intensities minus model fit) only. This P value quantifies the probability that MeDiChI will falsely report a peak with an intensity greater than or equal to that of the measured peak and is related to the peak's intensity relative to the noise across the dataset. Example peak detections for both CTCF and Scc/SA1 are shown, respectively, in Fig. 2 A and B, along with the MeDiChI-predicted peak location and intensity (green vertical line) and best fit to the data (red dashed curve).
Chromosome Preparation and Immunostaining.
Metaphase chromosomes were prepared from primary human fibroblasts treated with colcemid for 1 h. After hypotonic treatment in 0.075 M KCl for 10 min cells were spun onto slides in a cytocentrifuge. Chromosome preparations were incubated with CTCF antibody (Upstate Biotechnology, 06-917) for 1 h followed by incubation in a secondary FITC-labeled antibody (Vector). Cells were then fixed in 4% formaldehyde before counterstaining with Hoechst 33258. A minimum of 50 nuclei from three independent preparations of primary human fibroblasts was examined.
Supplementary Material
Acknowledgments.
We thank Charis Himeda for helpful advice on protein preparation for quantitative mass spectrometry, Stephen Tapscott and Sarah Mahoney (Fred Hutchinson Cancer Research Center, Seattle) for cell lines and information, William Schubach for critical reading and helpful comments, and the Institute for Systems Biology proteomics facility for technical support. This project has been funded in part by the National Heart, Lung, and Blood Institute Proteomics Initiative, National Institutes of Health, under contract no. N01-HV-28179 (to R.A.), and by National Institutes of Health Grants CA109597 (to A.K.), GM46883 (to C.M.D.), P50GM076547 and RGM077398A (to N.S.B and D.J.R.), and with National Science Foundation Grant BDI-0640950 (to N.S.B).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801273105/DCSupplemental.
References
- 1.Kosak ST, et al. Coordinate gene regulation during hematopoiesis is related to genomic organization. PLoS Biol. 2007;5:e309. doi: 10.1371/journal.pbio.0050309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Osborne CS, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 3.Wallace JA, Felsenfeld G. We gather together: Insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Capelson M, Corces VG. Boundary elements and nuclear organization. Biol Cell. 2004;96:617–629. doi: 10.1016/j.biolcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Krueger C, Osborne CS. Raising the curtains on interchromosomal interactions. Trends Genet. 2006;22:637–639. doi: 10.1016/j.tig.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 6.Yusufzai TM, et al. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 7.Filippova GN. Genetics and epigenetics of the multifunctional protein CTCF. Curr Top Dev Biol. 2008;80:337–360. doi: 10.1016/S0070-2153(07)80009-3. [DOI] [PubMed] [Google Scholar]
- 8.Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 9.Ishihara K, Oshimura M, Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell. 2006;23:733–742. doi: 10.1016/j.molcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 10.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filippova GN, et al. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev Cell. 2005;8:31–42. doi: 10.1016/j.devcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 13.Gombert WM, et al. The c-myc insulator element and matrix attachment regions define the c-myc chromosomal domain. Mol Cell Biol. 2003;23:9338–9348. doi: 10.1128/MCB.23.24.9338-9348.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ranish JA, Brand M, Aebersold R. Using stable isotope tagging and mass spectrometry to characterize protein complexes and to detect changes in their composition. Methods Mol Biol. 2007;359:17–35. doi: 10.1007/978-1-59745-255-7_2. [DOI] [PubMed] [Google Scholar]
- 15.Losada A. Cohesin regulation: Fashionable ways to wear a ring. Chromosoma. 2007;116:321–329. doi: 10.1007/s00412-007-0104-x. [DOI] [PubMed] [Google Scholar]
- 16.Sumara I, et al. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol. 2000;151:749–762. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reiss DJ, Facciotti MT, Baliga NS. Model-based deconvolution of genome-wide DNA binding. Bioinformatics. 2007;24:396–403. doi: 10.1093/bioinformatics/btm592. [DOI] [PubMed] [Google Scholar]
- 18.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 19.Stedman W, et al. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho DH, et al. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 21.Kmita M, Duboule D. Organizing axes in time and space; 25 years of colinear tinkering. Science. 2003;301:331–333. doi: 10.1126/science.1085753. [DOI] [PubMed] [Google Scholar]
- 22.Hauf S, et al. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waizenegger IC, et al. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- 24.Burke LJ, et al. CTCF binding and higher order chromatin structure of the H19 locus are maintained in mitotic chromatin. EMBO J. 2005;24:3291–3300. doi: 10.1038/sj.emboj.7600793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dorsett D. Roles of the sister chromatid cohesion apparatus in gene expression, development, and human syndromes. Chromosoma. 2007;116:1–13. doi: 10.1007/s00412-006-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donze D, et al. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rollins RA, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152:577–593. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rollins RA, et al. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol Cell Biol. 2004;24:3100–3111. doi: 10.1128/MCB.24.8.3100-3111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horsfield JA, et al. Cohesin-dependent regulation of Runx genes. Development. 2007;134:2639–2649. doi: 10.1242/dev.002485. [DOI] [PubMed] [Google Scholar]
- 30.Krantz ID, et al. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet. 2004;36:631–635. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tonkin ET, et al. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet. 2004;36:636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- 32.Deardorff MA, et al. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of Cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet. 2007;80:485–494. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Musio A, et al. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet. 2006;38:528–530. doi: 10.1038/ng1779. [DOI] [PubMed] [Google Scholar]
- 34.Ling JQ, et al. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 35.Splinter E, et al. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Z, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 37.Nasmyth K. How might cohesin hold sister chromatids together? Philos Trans R Soc Lond B. 2005;360:483–496. doi: 10.1098/rstb.2004.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gimenez-Abian JF, et al. Regulation of sister chromatid cohesion between chromosome arms. Curr Biol. 2004;14:1187–1193. doi: 10.1016/j.cub.2004.06.052. [DOI] [PubMed] [Google Scholar]
- 39.Parelho V, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 40.Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 41.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Himeda CL, et al. Quantitative proteomic identification of six4 as the trex-binding factor in the muscle creatine kinase enhancer. Mol Cell Biol. 2004;24:2132–2143. doi: 10.1128/MCB.24.5.2132-2143.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ranish JA, et al. Identification of TFB5, a new component of general transcription and DNA repair factor IIH. Nat Genet. 2004;36:707–713. doi: 10.1038/ng1385. [DOI] [PubMed] [Google Scholar]
- 44.Ranish JA, et al. The study of macromolecular complexes by quantitative proteomics. Nat Genet. 2003;33:349–355. doi: 10.1038/ng1101. [DOI] [PubMed] [Google Scholar]
- 45.Gygi SP, et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 46.O'Geen H, et al. Comparison of sample preparation methods for ChIP-chip assays. Biotechniques. 2006;41:577–580. doi: 10.2144/000112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.