Abstract
Mammalian spermatogenesis is a complex biological process that occurs within a highly organized tissue, the seminiferous epithelium. The coordinated maturation of spermatogonia, spermatocytes, and spermatids suggests the existence of precise programs of gene expression in these cells and in their neighboring somatic Sertoli cells. The objective of this study was to identify the genes that execute these programs. Rat seminiferous tubules at stages I, II–III, IV–V, VI, VIIa,b, VIIc,d, VIII, IX–XI, XII, and XIII–XIV of the cycle were isolated by microdissection, whereas Sertoli cells, spermatogonia plus early spermatocytes, pachytene spermatocytes, and round spermatids were purified from enzymatically dispersed testes. Microarray analysis by using Rat Genome 230 2.0 arrays identified 16,971 probe sets that recognized testicular transcripts, and 398 of these were identified as testis-specific. Expression of 1,286 probe sets were found to differ at least 4-fold between two cell types and also across the stages of the cycle. Pathway and annotated cluster analyses of those probe sets predicted that entire biological pathways and processes are regulated cyclically in specific cells. Important among these are the cell cycle, DNA repair, and embryonic neuron development. Taken together, these data indicate that stage-regulated gene expression is a widespread and fundamental characteristic of spermatogenic cells and Sertoli cells.
Keywords: array analysis, spermatogenesis, seminiferous tubules, stages of the cycle of the seminiferous epithellum, contraception
In mammals, spermatogenesis encompasses a series of precisely timed cellular events that take place in a highly organized tissue, the seminiferous epithelium. Within each cross-section of this tissue, spermatogonia, spermatocytes, and spermatids are in intimate physical association with somatic Sertoli cells, and, together, these cells progress synchronously through the stages of the cycle of the seminiferous epithelium (1, 2). Four cycles are required for spermatogonia, and their progeny to complete spermatogenesis. The synchrony of germ cell development has a significant effect on the structures and functions of both the germ cells and their associated somatic Sertoli cells, suggesting the existence of precise and coordinated cyclic programs of gene expression. However, because investigators have evaluated the stage-dependent changes in expression of only a few genes, whether or not cyclic gene expression is a rare or prevalent characteristic of these cells is unknown (3–7). It is also not known whether genes encoding components of important biological pathways and processes are coordinately regulated in a cyclic manner. However, such genes may represent new targets for the development of male contraceptives.
Given the coordinated nature of spermatogenesis, we hypothesized that the expression of large numbers of genes differ substantially, both between specific cell types within the seminiferous epithelium and within a given cell type as it progresses through the stages of the cycle. Therefore, we investigated global gene expression patterns in rat seminiferous tubules at defined stages of the cycle, in purified rat germ cells, and in Sertoli cells by using the Affymetrix GeneChip Rat Expression Array RAE230 2.0. This study detected 16,971 probe sets,** and analysis of their cell type and stage-regulated expression supported the conclusion that cyclic gene expression is a widespread and, therefore, fundamental characteristic of both spermatogenic and Sertoli cells. These data predicted that important biological pathways and processes are regulated as specific cell types progress through the stages of the cycle.
Results and Discussion
Characterization of the Testicular Transcriptome.
Analysis of the rat testicular transcriptome was accomplished by profiling seminiferous tubules at stages I, II–III, IV, V, VI, VIIa,b, VIIc,d, VIII, IX–XI, XII, and XIII–XIV, as well as purified Sertoli cells, spermatogonia plus early spermatocytes, pachytene spermatocytes and round spermatids (8–10). This approach identified 16,971 probe sets corresponding to ≈54% of those on the array. In contrast, only 9,888 probe sets were detected when RNA was isolated from whole testis. Table 1 shows the total number of probe sets expressed at each stage of the cycle and in each of the testicular cell types. The accuracy of array analysis was established by demonstrating that similar results were obtained when five different probe sets were quantified both by array analysis and by quantitative real time RT-PCR [supporting information (SI) Fig. S1].
Table 1.
Numbers of probe sets expressed at specific stages of the cycle and by individual cell types
| Sample analyzed | Total expressed | Specific to testis | Maximally expressed per stage or cell | Total stage-regulated or cell-regulated | No. of replicates |
|---|---|---|---|---|---|
| Stage | |||||
| Stage I | 7,885 | 225 | 116 | 947 | 5 |
| Stage II–III | 7,812 | 235 | 92 | 992 | 5 |
| Stage IV–V | 8,589 | 263 | 215 | 1,124 | 4 |
| Stage VI | 8,040 | 248 | 272 | 1,056 | 5 |
| Stage VIIa,b | 8,386 | 259 | 144 | 1,123 | 5 |
| Stage VIIc,d | 7,850 | 245 | 146 | 1,045 | 5 |
| Stage VIII | 8,364 | 245 | 147 | 1,042 | 5 |
| Stage IX–XI | 7,970 | 222 | 188 | 934 | 5 |
| Stage XII | 8,339 | 229 | 88 | 926 | 5 |
| Stage XIII–XIV | 9,888 | 240 | 204 | 929 | 5 |
| Total for all stages | 11,699 | 339 | 1612 | 1,612 | 49 |
| Cell type | |||||
| Spermatogonia plus early spermatocytes | 12,501 | 41 | 2910 | 6,665 | 3 |
| Pachytene spermatocytes | 9,102 | 278 | 1380 | 4,409 | 4 |
| Round spermatids | 8,026 | 303 | 2147 | 3,963 | 4 |
| Sertoli cells | 12,786 | 120 | 3113 | 699 | 4 |
| Total for all cell types | 16,608 | 370 | 9550 | 9,550 | 15 |
| Grand total | 16,971 | 398 | 10,005 | 64 |
To ascertain the percentage of the rat testis transcriptome that was testis-specific, we compared the gene expression data from these experiments to a proprietary database of microarray analysis of 21 other normal tissues. Results from this comparison indicate that 398 of 16,971 probe sets (2.3%) were testis-specific. Table 1 shows the number of testis-specific probe sets detected in tubules at defined stages of the cycle and in specific cell types. Dataset S1 lists the testis-specific probe sets, as well as their expression in testis and in 21 other organs.
To identify genes that are expressed in a testis-specific manner by both the mouse and the rat, the 346 mouse testis-specific probe sets previously identified by using the Affymetrix U74v2 array (supplemental tables 3 and 4 in ref. 11) were mapped to 103 probe sets on the rat RAE 230 2.0 array. Forty-two of these 103 probe sets were among the 398 testis-specific probe sets identified in our study and 27 of these map to known genes (Dataset S2).
Analysis of Defined Stages of the Cycle of the Seminiferous Epithelium.
Fourteen stages of the cycle have been identified in the rat; microarray analysis identified 11,699 expressed probe sets in the staged tubules (Table 1), and 1,612, 428, 42, and 13 probe sets were differentially expressed between any two stages or groups of stages by 4, 10, 50, and 100-fold, respectively. Dataset S3 lists the 42 probe sets whose expression vary at least 50-fold across the stages of the cycle. To gain a more complete understanding of the extent of stage-regulated gene expression, we chose to further characterize only the 1,612 probe sets that are differentially expressed by at least four-fold (P < 0.01). Table 1 lists the numbers of probe sets that are maximally expressed per stage, as well as the total number of stage-regulated probe sets expressed at each stage. This second set of numbers is larger because stage-regulated probe sets are expressed at multiple stages. Fig. S2 illustrates the stage-regulated expression of these probes sets, and Dataset S4 shows their Affymetrix probe set identifiers, gene description and symbol, and expression in each cell and at each stage.
Analysis of Specific Spermatogenic Cells and Sertoli Cells.
Microarray analysis of purified spermatogonia plus early spermatocytes, pachytene spermatocytes, round spermatids, and Sertoli cells identified 16,608 probe sets (Table 1). Data identified 14,113 probe sets, 9,550 probe sets, 771 probe sets, and 281 probe sets as differentially expressed 2-, 4-, 50-, and 100-fold or more, respectively, between any two cell types. Probe sets that were expressed 4-fold higher in one cell type compared with any of the other three cell types (P < 0.01) were selected for further analysis. Fig. S3 illustrates the relative expression patterns of the 9,550 probes sets differentially expressed between the different cell types.
Stage-Regulated Gene Expression in Spermatogonia, Spermatocytes, Spermatids, and Sertoli Cells.
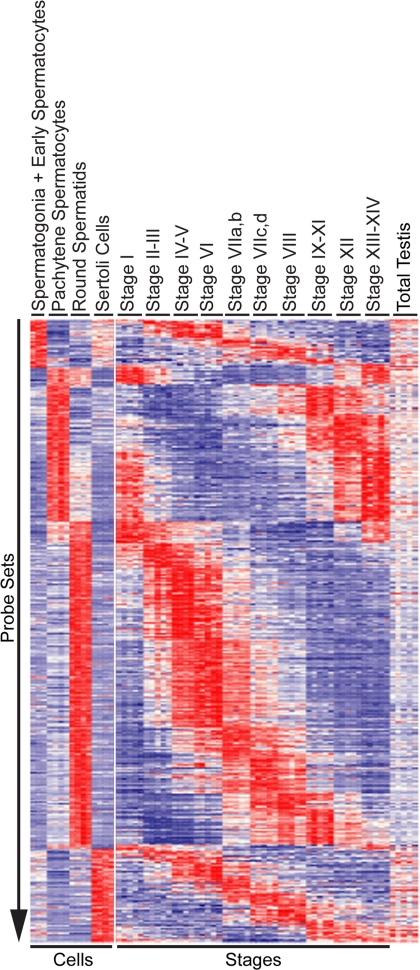
To identify probe sets whose expression differed both between cells and across the stages of the cycle, we intersected the 9,550 probe sets that were differentially expressed at least 4-fold between the testicular cell types with the 1,612 probe sets regulated at least 4-fold across the stages. This analysis identified 1,286 probe sets that recognized 98, 318, 677, and 193 stage-regulated transcripts that were maximally expressed in spermatogonia plus early spermatocytes, pachytene spermatocytes, round spermatids, and Sertoli cells, respectively. Fig. 1 shows a heat map of the expression of all 1,286 probe sets ordered first by the testicular cell type exhibiting maximally expression and second by the stage or group of stages exhibiting maximal expression. Different cell types exhibit distinct patterns of stage-regulated gene expression. Spermatogonia plus early spermatocytes exhibit elevated probe set expression at stages II–XI, pachytene spermatocytes exhibit elevated probe set expression at stages IX–XIV, and round spermatids exhibit elevated probe set expressed at stages IV–VI. Sertoli cells maximally expressed distinct probe sets at each of the stages in the cycle.
Fig. 1.
Relative expression levels of 1,286 probe sets 4-fold regulated in both testicular cell types and during the cycle of the seminiferous epithelium. These probe sets were ordered by cell type and by stage with the highest expression. Data were visualized in Spotfire 8.0. Each probe set was z-scored normalized and color-coded (red = high expression, white = median expression, blue = low expression).
Predicted Regulation of Biological Pathways in Specific Cell Types.
To identify regulated pathways in spermatogenic or Sertoli cells, probe sets whose expression differed at least 4-fold between at least two cell types were sorted by the cell with the highest level of expression. These lists were then analyzed to identify both the genes encoding those probe sets and the biological pathways that were predicted by the gene expression profiles to be up-regulated in each cell type. Pathways whose activities potentially varied with progression of the stages of the cycle were defined as those that included at least two genes whose expression varied at least 4-fold across the stages. This analysis identified four, one, and three stage-regulated pathways in spermatids, spermatocytes, and Sertoli cells, respectively (Table 2). Dataset S5 lists the genes in these pathways. No stage-regulated pathways were identified in spermatogonia plus early spermatocytes.
Table 2.
Identification of pathways predicted to be regulated in a stage-dependent manner in spermatids, spermatocytes, and Sertoli cells
| Kegg pathway | No. of maximum expressed genes/pathway | No. of stage-regulated genes per pathway |
|---|---|---|
| Spermatid | ||
| DNA polymerase | 7 | 2 |
| Cell cycle | 15 | 5 |
| Insulin signaling | 17 | 4 |
| Purine metabolism | 15 | 6 |
| Spermatocyte | ||
| Insulin signaling | 11 | 2 |
| Sertoli cell | ||
| TGF-β signaling pathway | 20 | 2 |
| Focal adhesion | 42 | 2 |
| Fatty acid metabolism | 12 | 2 |
Four potential stage-regulated pathways were identified in spermatids. One pathway mediates responses to extracellular signals, two regulate DNA or cell replication, and one is involved in purine metabolism. In spermatocytes, the one stage-regulated pathway controls responses to extracellular signals. This analysis also identified three stage-regulated pathways in Sertoli cells. Consistent with their function as organizing the seminiferous epithelium, one pathway regulates cell–cell adhesion and another mediates signaling by members of the TGFβ family of growth factors. It is noteworthy that as these somatic cells define the physiological environment within the seminiferous tubule, fatty acid metabolism was also identified as stage-regulated.
Predicted Stage-Dependent Regulation of the Cell Cycle in Spermatocytes and Spermatids.
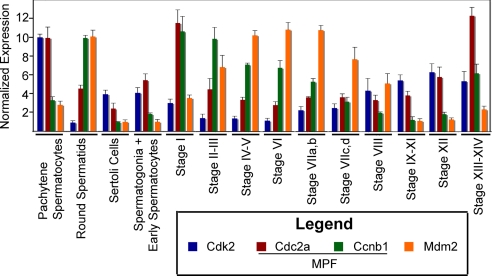
Given the fundamental importance of the meiotic cell cycle to spermatogenesis, we reanalyzed the data to define all genes with the annotation of cell cycle (Gene Ontology accession no. GO:0007049) whose expression differed significantly both between cell types and between stages of the cycle regardless of the fold difference in expression (P < 0.01). Cell cycle genes were highly over-represented in these data (P < 1 × 10−9), and 83 and 71 cell cycle genes exhibited elevated expression in pachytene spermatocytes and round spermatids, respectively (Table 3). Forty-seven of the cell cycle genes enriched in spermatocytes were maximally expressed at stages XII or XIII–XIV and 20 genes at stage I. Increased expression at these stages is noteworthy because spermatocytes complete their long meiotic prophase at stages XII–XIII, undergo the two meiotic cell divisions at stage XIV, and produce haploid spermatids at stage I (12). Examination of individual genes shows that expression of some cell cycle genes begin to increase at stages VIIa,b to VIII. An example is cyclin-dependent kinase 2 (Cdk2), which is required for completion of the prophase of meiosis I (13) (Fig. 2). Our observation that some cell cycle genes are transcriptionally expressed at higher levels in purified round spermatids than in purified pachytene spermatocytes is consistent with previous reports (14). However, 18 of these genes were maximally expressed at stages XIII–XIV or stage I (Table 3), indicating that their increased expression begins in spermatocytes. Of particular note is the 3.4-fold increase in expression of cyclin B1 (Ccnb1) from stage XII to stages XII–XIV that occurs coincidently with a doubling in expression of Cdc2a (cell division cycle 2 homolog A) (Fig. 2.). Because Ccnb1 and Cdc2a encode the two subunits of maturation-promoting factor (MPF), we suggest that this coordinated transcriptional expression ensures that there is sufficient MPF to drive spermatocytes through the G2/M transition of meiosis.
Table 3.
Number of genes involved in cell cycle regulation and DNA repair that are maximally expressed in a given cell type at a given stage
| Pathway or process | Cell type with maximal expression | Stage of maximal expression |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II-III | IV-V | VI | VIIa,b | VIIc,d | VIII | IX-XI | XII | XIII-XIV | Σ | ||
| Cell cycle | Pachytene spermatocytes | 20 | 1 | 0 | 0 | 0 | 2 | 5 | 8 | 18 | 29 | 83 |
| Cell cycle | Round spermatids | 15 | 4 | 13 | 14 | 1 | 6 | 4 | 11 | 0 | 3 | 71 |
| DNA repair | Pachytene spermatocytes | 3 | 1 | 0 | 0 | 0 | 1 | 1 | 7 | 3 | 14 | 30 |
| DNA repair | Round spermatids | 3 | 2 | 3 | 7 | 1 | 3 | 0 | 1 | 0 | 2 | 22 |
Fig. 2.
Transcriptional expression of cell cycle genes by pachytene spermatocytes, round spermatids, Sertoli cells, spermatogonia plus early spermatocytes, and by seminiferous tubules at defined stages of the cycle. In this figure and in Figs. 3 and 4, the testicular cell-type with the highest level of expression was defined as having a mean level of expression of 10 units. Data for the other cell-types and for staged tubules were normalized accordingly. Cdk2 and Ccnb1 encode the subunits of MPF. Data are expressed as the mean + SD. The stages of the cycle are defined by germ cell morphology and different stages persist for different but precise periods of time (12, 46).
Table 3 indicates that at stages IV–VI, spermatids also display an increased expression of 27 cell cycle genes. Eight of those genes are categorized by the Gene Ontology biological processes GO:0008285 (negative regulation of cell proliferation) and/or GO:0007050 (cell cycle arrest). Fig. 2 shows the expression of one of these genes, Mdm2 (transformed mouse 3T3 cell double minute 2). Thus, with completion of the meiotic cell cycle, there is increased expression in spermatids of genes whose products repress the cell cycle machinery.
Predicted Stage-Dependent Regulation of DNA Repair in Spermatocytes and Spermatids.
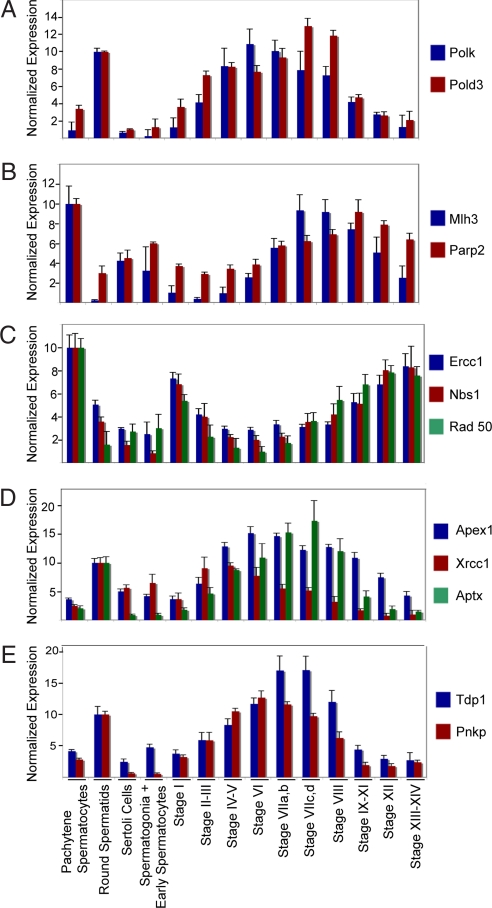
Transcripts encoding seven DNA polymerases or their subunits are expressed at least 4-fold higher in spermatids than in any other testicular cell type and two of these, Polk (DNA-directed polymerase κ) and the Pold3 (accessory subunit for Pol δ), are expressed in a stage-regulated manner (Fig. 3A). Their regulated expression was noteworthy because spermatids have enhanced DNA repair capacity and because Polk and Pold3 participate in nucleotide excision repair and base excision repair, respectively (15–17).
Fig. 3.
Transcriptional expression of genes involved in base excision repair, nucleotide excision repair, recombinational repair, and/or single-strand break repair by pachytene spermatocytes, round spermatids, Sertoli cells, spermatogonia and early spermatocytes, and seminiferous tubules at defined stages of the cycle of the seminiferous epithelium. Shown are messenger RNA levels for Polk, Pold3 (A); Mlh3, Parp2 (B); Ercc1, Nbs1, Rad50 (C); Apex1, Xrcc1, Aptx (D); and Tdp1 and Pnpk (E) as determined by microarray analysis. Data are expressed as the mean + SD.
Because preservation of genome integrity is required for male fertility, we sought to gain as complete a picture as possible of the regulation of DNA repair processes in spermatogenic cells. To that end, we identified all probe sets with the GO annotation, GO:0006281 (DNA repair), whose expressions varied significantly between cells and between stages (P < 0.01), regardless of the fold change in expression of their cognate transcripts. This reanalysis identified 274 probe sets with this annotation, and these were significantly overrepresented in the data (P < 0.001). This reanalysis revealed 30 and 22 regulated DNA repair genes that are maximally expressed in isolated spermatocytes and spermatids, respectively. Table 3, which shows the numbers of DNA repair genes maximally expressed by spermatocytes and by spermatids at each stage of the cycle, suggests that there are waves of expression of DNA repair genes in these cells.
One of the first DNA repair genes to show increased expression in pachytene spermatocytes is Mlh3 (MutL homolog 3), which increases 24-fold from its minimum stages II–III to its maximum at stage VIIc,d. (Fig. 3B). This increase correlates with the accumulation of Mlh3 protein in recombination nodules formed during synapses of homologous chromosomes (18). In pachytene spermatocytes, Mlh3 protein is bound to the Msh4 protein. Because Msh4 is involved in resolving Holiday junctions, the function of Mlh3 may extend beyond its well established role in mismatch DNA repair to the repair of DNA double-strand breaks formed during homologous recombination (19, 20). The stage-regulated increase in expression of Mlh3 is accompanied by increased expression of Parp2 [poly(ADP-ribose) polymerase family, member 2], which is involved in base excision repair (21) (Fig. 3B). This increase is of note because Parp2 is required for efficient meiosis and spermiogenesis (21). Ercc1 (excision repair cross-complementing rodent repair deficiency, complementation group 1), Nbs1 (Nijmegen breakage syndrome 1), and Rad50 (Rad 50 homolog Saccharomyces cerevisiae), which encode components of the recombination repair pathway, reveal a third wave of DNA repair gene expression by pachytene spermatocytes. The expression of these genes, which is lowest at stage IV to VI, is increased 3- to 8-fold by stages XIII–XIV (Fig. 3C). These increases are important because Nbs1 and Rad50 are part of the MRE11 complex that is required for yeast to generate double-strand breaks during meiotic recombination and the numbers of double-strand breaks are increased in Ercc1-null mouse spermatogenic cells (22, 23). This suggests that Ercc1 acts after Nbs1 and Rad 50 during meiotic chromosome homologous recombination. With completion of meiosis, there is decreased expression of genes involved in meiotic recombination and increased expression of genes involved in base excision repair and/or single-strand break repair. Our data predict that spermatids come primed for the first step in base excision repair, removal of the damaged base, because expression of genes encoding Mpg (N-methylpurine-DNA glycosylase), Nthl1 (thymine glycol DNA glycosylase/AP lyase), and Ogg1 (8-oxoguanine DNA-glycosylase) are elevated in both pachytene spermatocytes and round spermatids (data not shown). Increased transcriptional expression of apurinic/apyrimidinic endonuclease 1 (Apex1), the endonuclease that makes the single-stand cut 5′ to the abasic site, begins at stages IV–V in spermatids (Fig. 3D). This occurs coordinately with increased expression of Xrcc1 (x-ray repair cross-complementing group 1), the scaffold for the assembly of the enzymes catalyzing the remaining steps in base excision repair (24) (Fig. 3D). Xrcc1 also forms a complex with Aptx (aprataxin) and Lig3 (ligase III, DNA, ATP-dependent) to initiate single-strand break repair (25) and transcriptional expression of Xrcc1 and Aptx overlap (Fig. 3D). As with the DNA glycosylases, transcriptional expression of Lig3 is increased in spermatocytes and persists in spermatids (data not shown). Transcriptional expression of two other enzymes involved in single-strand break repair, Tdp1 (tyrosyl-DNA phosphodiesterase 1) and Pnkp (polynucleotide kinase 3′ phosphatase), are up-regulated in spermatids coincidently with Aptx (Fig. 3E) (26–28).
Stage-Dependent Expression by Sertoli Cells of Genes Previously Shown to Regulate Embryonic Neuron Development.
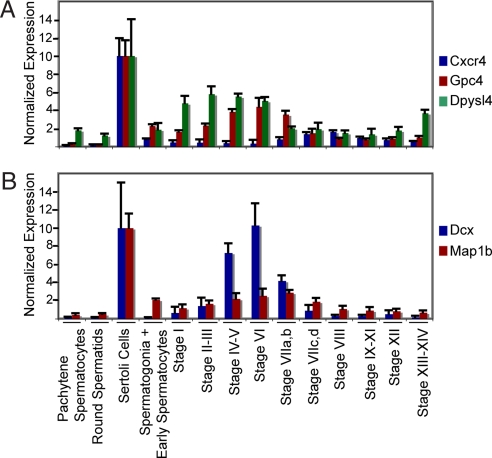
Sertoli cells define the environment in which spermatogenesis occurs and move the developing male gametes from the basement membrane to the lumen of the seminiferous tubule (29). Thus, we hypothesized that the genes encoding these processes are expressed at significantly higher levels by Sertoli cells than by any spermatogenic cell. To identify these genes, we focused on the 81 stage-regulated probe sets that were expressed at least 3-fold higher in mature Sertoli cells than in spermatogonia, spermatocytes, or spermatids. Functional annotation clustering sorted these probe sets into six clusters, with a significant enrichment of genes described by at least one Uniprot keyword or GOterm (Dataset S6). Genes in these clusters encoded secretory products, glycoproteins, proteases, cell surface receptors, and lipid -metabolizing enzymes. Additionally, seven genes were identified that were categorized by the gene ontology biological process GO:0007399 (nervous system development) (P < 0.04). Three of these genes encode components of signal transduction pathways that regulate the movement and/or development of neurons within the embryonic brain: chemokine (C-X-C motif) receptor 4 (Cxcr4), glypican 4 (Gpc4), and dihydropyrimidinase-like 4 (Dpysl4) (30–35). For example, Cxcr4 regulates the migration of embryonic neurons into the cerebellum (35). The patterns of stage-regulated expression of these genes by mature Sertoli cells differ because Cxcr4, Gpc4, and Dpsl are maximally expressed at stages II–III, VI, and VIII, respectively (Fig. 4A). Other genes in these pathways are also expressed in the seminiferous epithelium. For example, stromal cell line-derived factor 1, the ligand for the cell surface receptor, Cxcr4, is expressed by spermatogonia and early spermatocytes (Fig. S4). Gpc4 is a GPI-anchored heparin sulfate proteoglycan that binds to growth factors, including FGF-2, and thereby modulates their effects on target cells (31, 36). FGF-2 is produced by pachytene spermatocytes (37). Dpysl4 transduces the signal that is generated by the binding of the semaphorin family of signaling molecules to a heterodimeric cell surface receptor encoded by a member of the plexin family and by RhoA (33). Not only are plexin B2 and RhoA mRNA detected in Sertoli cells, but semaphorin 4A is expressed in a stage-dependent manner by spermatids (Fig. S4).
Fig. 4.
Transcriptional expression by Sertoli cells and by spermatogenic cells of genes previously shown to regulate embryonic neuron development. Shown are messenger RNA levels for Cxcr4, Gpc4, Dpysl4 (A) and Dcx and Map1b (B) as determined by microarray analysis. Data are expressed as the mean + SD.
Two other genes in this cluster encode the microtubule associated proteins, doublecortin (Dcx) and microtubule-associated protein 1b (Map1b), that play important roles in neuronal guidance in the embryonic brain (38, 39). In mature Sertoli cells, Dcx mRNA concentration increases 18-fold from stage I to stage VI and then decreases 60% at stage VIIa,b. In contrast, Map1b expression increases gradually from stage I and reaches its maximum at stage VIIa,b (Fig. 4B). In Sertoli cells, the functions of the proteins encoded by these two genes are undoubtedly modulated by phosphorylation, which, in other cell types, decreases their ability to stabilize microtubules (40). Sertoli cells express Cdk-5, which phosphorylates doublecortin (Fig. S4). Sertoli cells also express TrkA and p75, the heterodimeric receptor for NGF that regulates phosphorylation of Map1b and round spermatids and pachytene spermatocytes express NGF (41–43).
These data support the conclusion that there are multiple stage-regulated pathways in Sertoli cells that regulate cell migration. However, mature Sertoli cells are anchored to a basement membrane. What then might be the functions of cell migration pathways in these cells? Cell movement requires marked changes in cell shape, volume, cytoskeleton organization, and interaction with other cells. It is, therefore, noteworthy that there are marked stage-regulated changes in the volume and surface area of Sertoli cells, as well as in the amount of Sertoli cell plasma membrane in contact with spermatogonia, spermatocytes, and spermatids (37, 44). Therefore, although the Sertoli cells do not themselves move, the changes that they undergo are consistent with their role in moving germ cells from the basement membrane to the lumen of the seminiferous tubule.
Summary and Conclusions.
This study has evaluated both cell-specific and stage-regulated gene expression by spermatogenic cells and Sertoli cells. Three hundred ninety-six testis-specific probe sets have been identified, and some of these may be potential contraceptive targets. Furthermore, our results show that stage-regulated gene expression is a widespread and fundamental characteristic of spermatocytes, spermatids, and Sertoli cells. These results predict that activities of biological pathways and processes are precisely regulated in these cells as a consequence of both cell-specific and stage-regulated gene expression. Our data demonstrate that late in meiosis, there is a coordinated increase in the expression of genes needed for the completion of the meiotic cell cycle and for the repair of double-strand breaks in DNA, a requirement for homologous recombination. Upon completion of meiosis, expression of those genes declines and expression of genes required for cell cycle repression and base excision and single-strand DNA break repair increases. Furthermore, our data suggest that biological processes that drive nerve cell migration in the developing brain also function in the adult Sertoli cell to direct germ cells to specific locations within the seminiferous epithelium. Therefore, taken together, these data support the conclusion that stage-regulated gene expression is an essential part of the regulatory program that controls the orderly and coordinated development of male germ cells.
Methods
Tissue and Cell Collection.
Seminiferous tubules, Sertoli cells, pachytene spermatocytes, and round spermatids were isolated from fifty-five 60-day-old Sprague–Dawley rats, and a pool of spermatogonia and early spermatocytes was isolated from eighteen 20-day-old rats. The caveat inherent in isolating spermatogonia plus early spermatocytes from immature rats is that the gene expression profile of these cells is the same in immature and mature animals. The Johns Hopkins University Animal Care and Use Committee approved these experiments.
Ten centimeters of seminiferous tubules at the following stages or groups of stages were isolated by transillumination-assisted microdissection (8): I, II–III, IV–V, VI, VIIa,b, VIIc,d, VIII, IX–XI, XII, and XIII–XIV. Cells were isolated from enzyme-dispersed testes, and mature Sertoli cells were obtained at ≈85% purity (10). Round spermatids, pachytene spermatocytes, and spermatogonia plus early spermatocytes were isolated by centrifugal elutriation (9) (see SI Materials and Methods and Table S1 for details). RNA was isolated and quantified, and its quality determined as previously described (8). The number of independent cell isolates that were analyzed is shown in Table 1.
Microarray Processing.
RNA from staged tubules or purified testicular cell samples was isolated, labeled, and hybridized to RAE230 2.0 arrays (Affymetrix), and the data were analyzed as previously described (45). A probe set initially was defined as differentially expressed if the difference between two groups within the comparison met the following criteria: (i) the probe set was detected in at least one of the experimental groups (signal intensity ≥50 units); (ii) there was at least a 4-fold difference in expression between at least two groups; and (iii) the P value based on an ANOVA test was ≤0.01. After this analysis, data were reanalyzed to identify all probe sets that were significantly (P < 0.01) differentially expressed between cell types and between stages, independent of the fold change in expression. Pathway and annotated cluster analysis was performed by using the functional annotation tools of David Bioinformatics Resources (http://david.abcc.ncifcrf.gov).
Tissue Specificity.
Probe sets recognizing transcripts expressed at a given stage, group of stages, or in a purified testicular cell were screened for tissue selectivity by comparison with a Wyeth proprietary database of RAE230 profiling data from 21 normal rat tissues. Each tissue was analyzed three to 12 times. Data were normalized to a mean signal intensity value of 100 in GCOS (Affymetrix). Probe sets were considered testis-selective if the following conditions were met: (i) the probe set had a 67% present (P) call in at least one staged sample or purified testicular cell sample; (ii) the expression of the probe set in the testis was ≥50 signal units; (iii) the probe set was never detected (0% P call) in any of the 21 other tissues; and (iv) the signal for a probe sets was <50 signal units in all 21 tissues.
Supplementary Material
Acknowledgments.
This research was supported in part by National Institutes of Health Grant 1 R01 HD044183 (to W.W.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE8978).
For brevity, we define a probe set as a transcript that binds to a set of oligonucleotides on the array that are complementary to a specific nucleotide sequence.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709854105/DCSupplemental.
References
- 1.Parvinen M, Vihko KK, Toppari J. Cell interactions during the seminiferous epithelial cycle. Int Rev Cytol. 1986;104:115–151. doi: 10.1016/s0074-7696(08)61925-7. [DOI] [PubMed] [Google Scholar]
- 2.Wing TY, Christensen AK. Morphometric studies on rat seminiferous tubules. Am J Anat. 1982;165:13–25. doi: 10.1002/aja.1001650103. [DOI] [PubMed] [Google Scholar]
- 3.Charron M, Folmer JS, Wright WW. A 3-kilobase region derived from the rat cathepsin L gene directs in vivo expression of a reporter gene in sertoli cells in a manner comparable to that of the endogenous gene. Biol Reprod. 2003;68:1641–1648. doi: 10.1095/biolreprod.102.011619. [DOI] [PubMed] [Google Scholar]
- 4.Mali P, et al. Localization of protamine 1 mRNA in different stages of the cycle of the rat seminiferous epithelium. J Cell Biol. 1988;107:407–412. doi: 10.1083/jcb.107.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kistler WS, Henriksen K, Mali P, Parvinen M. Sequential expression of nucleoproteins during rat spermiogenesis. Exp Cell Res. 1996;225:374–381. doi: 10.1006/excr.1996.0188. [DOI] [PubMed] [Google Scholar]
- 6.Morales C, Hugly S, Griswold MD. Stage-dependent levels of specific mRNA transcripts in Sertoli cells. Biol Reprod. 1987;36:1035–1046. doi: 10.1095/biolreprod36.4.1035. [DOI] [PubMed] [Google Scholar]
- 7.Morales CR, Alcivar AA, Hecht NB, Griswold MD. Specific mRNAs in Sertoli and germinal cells of testes from stage synchronized rats. Mol Endocrinol. 1989;3:725–733. doi: 10.1210/mend-3-4-725. [DOI] [PubMed] [Google Scholar]
- 8.Kotaja N, et al. Preparation, isolation and characterization of stage-specific spermatogenic cells for cellular and molecular analysis. Nat Methods. 2004;1:249–254. doi: 10.1038/nmeth1204-249. [DOI] [PubMed] [Google Scholar]
- 9.Shaper NL, Wright WW, Shaper JH. Murine β1,4-galactosyltransferase: Both the amounts and structure of the mRNA are regulated during spermatogenesis. Proc Natl Acad Sci USA. 1990;87:791–795. doi: 10.1073/pnas.87.2.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anway MD, Folmer J, Wright WW, Zirkin BR. Isolation of Sertoli cells from adult rat testes: An approach to ex vivo studies of sertoli cell function. Biol Reprod. 2003;68:996–1002. doi: 10.1095/biolreprod.102.008045. [DOI] [PubMed] [Google Scholar]
- 11.Schultz N, Hamra FK, Garbers DL. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc Natl Acad Sci USA. 2003;100:12201–12206. doi: 10.1073/pnas.1635054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leblond CP, Clermont Y. Definition of the stages of the cycle of the seminiferous epithelium in the rat. Ann N Y Acad Sci. 1952;55:548–573. doi: 10.1111/j.1749-6632.1952.tb26576.x. [DOI] [PubMed] [Google Scholar]
- 13.Ortega S, et al. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat Genet. 2003;35:25–31. doi: 10.1038/ng1232. [DOI] [PubMed] [Google Scholar]
- 14.Gromoll J, Wessels J, Rosiepen G, Brinkworth MH, Weinbauer GF. Expression of mitotic cyclin B1 is not confined to proliferating cells in the rat testis. Biol Reprod. 1997;57:1312–1319. doi: 10.1095/biolreprod57.6.1312. [DOI] [PubMed] [Google Scholar]
- 15.Intano GW, et al. Base excision repair is limited by different proteins in male germ cell nuclear extracts prepared from young and old mice. Mol Cell Biol. 2002;22:2410–2418. doi: 10.1128/MCB.22.7.2410-2418.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogi T, Lehmann AR. The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nat Cell Biol. 2006;8:640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- 17.Stucki M, et al. Mammalian base excision repair by DNA polymerases delta and epsilon. Oncogene. 1998;17:835–843. doi: 10.1038/sj.onc.1202001. [DOI] [PubMed] [Google Scholar]
- 18.Lipkin SM, et al. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat Genet. 2002;31:385–390. doi: 10.1038/ng931. [DOI] [PubMed] [Google Scholar]
- 19.Moens PB, Marcon E, Shore JS, Kochakpour N, Spyropoulos B. Initiation and resolution of interhomolog connections: Crossover and non-crossover sites along mouse synaptonemal complexes. J Cell Sci. 2007;120:1017–1027. doi: 10.1242/jcs.03394. [DOI] [PubMed] [Google Scholar]
- 20.Santucci-Darmanin S, et al. The DNA mismatch-repair MLH3 protein interacts with MSH4 in meiotic cells, supporting a role for this MutL homolog in mammalian meiotic recombination. Hum Mol Genet. 2002;11:1697–1706. doi: 10.1093/hmg/11.15.1697. [DOI] [PubMed] [Google Scholar]
- 21.Dantzer F, et al. Poly(ADP-ribose) polymerase-2 contributes to the fidelity of male meiosis I and spermiogenesis. Proc Natl Acad Sci USA. 2006;103:14854–14859. doi: 10.1073/pnas.0604252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farah JA, Cromie G, Steiner WW, Smith GR. A novel recombination pathway initiated by the Mre11/Rad50/Nbs1 complex eliminates palindromes during meiosis in Schizosaccharomyces pombe. Genetics. 2005;169:1261–1274. doi: 10.1534/genetics.104.037515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johzuka K, Ogawa H. Interaction of Mre11 and Rad50: Two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics. 1995;139:1521–1532. doi: 10.1093/genetics/139.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair. 2003;2:955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 25.Luo H, et al. A new XRCC1-containing complex and its role in cellular survival of methyl methanesulfonate treatment. Mol Cell Biol. 2004;24:8356–8365. doi: 10.1128/MCB.24.19.8356-8365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jilani A, et al. Molecular cloning of the human gene, PNKP, encoding a polynucleotide kinase 3′-phosphatase and evidence for its role in repair of DNA strand breaks caused by oxidative damage. J Biol Chem. 1999;274:24176–24186. doi: 10.1074/jbc.274.34.24176. [DOI] [PubMed] [Google Scholar]
- 27.El-Khamisy SF, Caldecott KW. DNA single-strand break repair and spinocerebellar ataxia with axonal neuropathy-1. Neuroscience. 2007;145:1260–1266. doi: 10.1016/j.neuroscience.2006.08.048. [DOI] [PubMed] [Google Scholar]
- 28.Nitiss KC, Malik M, He X, White SW, Nitiss JL. Tyrosyl-DNA phosphodiesterase (Tdp1) participates in the repair of Top2-mediated DNA damage. Proc Natl Acad Sci USA. 2006;103:8953–8958. doi: 10.1073/pnas.0603455103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mruk DD, Cheng CY. Sertoli-Sertoli and Sertoli-germ cell interactions and their significance in germ cell movement in the seminiferous epithelium during spermatogenesis. Endocr Rev. 2004;25:747–806. doi: 10.1210/er.2003-0022. [DOI] [PubMed] [Google Scholar]
- 30.Liapi A, et al. Stromal-derived factor 1 signalling regulates radial and tangential migration in the developing cerebral cortex. Dev Neurosci. 2008;30:117–131. doi: 10.1159/000109857. [DOI] [PubMed] [Google Scholar]
- 31.Hagihara K, Watanabe K, Chun J, Yamaguchi Y. Glypican-4 is an FGF2-binding heparan sulfate proteoglycan expressed in neural precursor cells. Dev Dyn. 2000;219:353–367. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1059>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 32.Ohkawara B, Yamamoto TS, Tada M, Ueno N. Role of glypican 4 in the regulation of convergent extension movements during gastrulation in Xenopus laevis. Development. 2003;130:2129–2138. doi: 10.1242/dev.00435. [DOI] [PubMed] [Google Scholar]
- 33.Quinn CC, Gray GE, Hockfield S. A family of proteins implicated in axon guidance and outgrowth. J Neurobiol. 1999;41:158–164. [PubMed] [Google Scholar]
- 34.Alabed YZ, Pool M, Ong Tone S, Fournier AE. Identification of CRMP4 as a convergent regulator of axon outgrowth inhibition. J Neurosci. 2007;27:1702–1711. doi: 10.1523/JNEUROSCI.5055-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou YR, Kottmann AH, Kuroda M, Taniuchi I, Littman DR. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 1998;393:595–599. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]
- 36.Fico A, Maina F, Dono R. Fine-tuning of cell signalling by glypicans. Cell Mol Life Sci. 2008 doi: 10.1007/s00018-007-7471-6. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Franca LR, Ghosh S, Ye SJ, Russell LD. Surface and surface-to-volume relationships of the Sertoli cell during the cycle of the seminiferous epithelium in the rat. Biol Reprod. 1993;49:1215–1228. doi: 10.1095/biolreprod49.6.1215. [DOI] [PubMed] [Google Scholar]
- 38.Koizumi H, et al. Doublecortin maintains bipolar shape and nuclear translocation during migration in the adult forebrain. Nat Neurosci. 2006;9:779–786. doi: 10.1038/nn1704. [DOI] [PubMed] [Google Scholar]
- 39.Meixner A, et al. MAP1B is required for axon guidance and Is involved in the development of the central and peripheral nervous system. J Cell Biol. 2000;151:1169–1178. doi: 10.1083/jcb.151.6.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Onishi M, Yasunaga T, Tanaka H, Nishimune Y, Nozaki M. Gene structure and evolution of testicular haploid germ cell-specific genes, Oxct2a and Oxct2b. Genomics. 2004;83:647–657. doi: 10.1016/j.ygeno.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 41.Session DR, et al. Cyclin-dependent kinase 5 is expressed in both Sertoli cells and metaphase spermatocytes. Fertil Steril. 2001;75:669–673. doi: 10.1016/s0015-0282(00)01794-5. [DOI] [PubMed] [Google Scholar]
- 42.Goold RG, Gordon-Weeks PR. NGF activates the phosphorylation of MAP1B by GSK3β through the TrkA receptor and not the p75(NTR) receptor. J Neurochem. 2003;87:935–946. doi: 10.1046/j.1471-4159.2003.02062.x. [DOI] [PubMed] [Google Scholar]
- 43.Parvinen M, et al. Expression of beta-nerve growth factor and its receptor in rat seminiferous epithelium: Specific function at the onset of meiosis. J Cell Biol. 1992;117:629–641. doi: 10.1083/jcb.117.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kerr JB. A light microscopic and morphometric analysis of the Sertoli cell during the spermatogenic cycle of the rat. Anat Embryol. 1988;177:341–348. doi: 10.1007/BF00315842. [DOI] [PubMed] [Google Scholar]
- 45.Jelinsky SA, et al. The rat epididymal transcriptome: Comparison of segmental gene expression in the rat and mouse epididymides. Biol Reprod. 2007;76:561–570. doi: 10.1095/biolreprod.106.057323. [DOI] [PubMed] [Google Scholar]
- 46.Clermont Y, Harvey SC. Duration of the cycle of the seminiferous epithelium of normal, hypophysectomized and hypophysectomized-hormone treated albino rats. Endocrinology. 1965;76:80–89. doi: 10.1210/endo-76-1-80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.