Abstract
As paleopolyploid genomes evolve, the expression profiles of retained gene pairs are expected to diverge. To examine this divergence process on a large scale in a vertebrate system, we compare Xenopus laevis, which has retained ≈40% of loci in duplicate after a recent whole-genome duplication (WGD), with its unduplicated relative Silurana (Xenopus) tropicalis. This comparison of ingroup pairs to an outgroup allows the direction of change in expression profiles to be inferred for a set of 1,300 X. laevis pairs, relative to their single orthologs in S. tropicalis, across 11 tissues. We identify 68 pairs in which X. laevis is inferred to have undergone a significant reduction of expression in at least two tissues since WGD. Of these pairs, one-third show evidence of subfunctionalization, with decreases in the expression levels of different gene copies in two different tissues. Surprisingly, we find that genes with slow rates of evolution are particularly prone to subfunctionalization, even when the tendency for highly expressed genes to evolve slowly is controlled for. We interpret this result to be an effect of allopolyploidization. We then compare the outcomes of this WGD with an independent one that happened in the teleost fish lineage. We find that if a gene pair was retained in duplicate in X. laevis, the orthologous pair is more likely to have been retained in duplicate in zebrafish, suggesting that similar factors, among them subfunctionalization, determined which gene pairs survived in duplicate after the two WGDs.
Keywords: rate of evolution, Silurana tropicalis, whole-genome duplication
Polyploidy, also termed whole-genome duplication (WGD) is a frequent phenomenon in eukaryotes (1). A WGD is followed by extensive and rapid genome restructuring involving many gene losses, so that only one of the two gene copies remains in most genomes that underwent ancient polyploidization [for example, fish and yeast (2, 3)]. Alterations in function are expected among genes retained in duplicate. In some cases, one copy may acquire a new function (neofunctionalization), while the other keeps the ancestral function. The models of Lynch and Force (4, 5) also propose the existence of subfunctionalization, in which each copy retains a subset of the functions of the ancestral gene. Sub and neofunctionalization models make different predictions about the rate and symmetry of sequence evolution in the duplicates.
Asymmetry in evolutionary rates between the protein sequences of the two copies is often interpreted as a footprint of neofunctionalization, especially if it is associated with evidence of positive selection in the accelerated copy (6). Several studies of paleopolyploid genomes have shown that rate asymmetry between the two copies can be widespread. For example, asymmetry was seen in 6% of retained gene pairs in Xenopus laevis and in 25–36% of pairs in teleost fishes (6–8). Relatively few examples of subfunctionalization of duplicated genes have been demonstrated so far, the best-known being those of fish mitf (9), sox9 (10), synapsin (11), POMC (12), mbx (13), and the plant gene RPL32-SODcp (14). A few studies have attempted to detect subfunctionalization on a larger scale after WGD. Aury et al. (15) used successive rounds of WGD in Paramecium to test Force et al.'s (5) prediction that subfunctionalized gene pairs should be resistant to reduplication. Their results suggest that subfunctionalization has occurred, but only rarely, in Paramecium genes. Other studies of subfunctionalization after WGD have focused on complementary amino acid substitution in protein pairs (6) and on the differential loss of regulatory regions between duplicated copies of developmental genes (16).
The most powerful method currently available to study the divergence of function between duplicated genes on a large scale is the analysis of their transcription profiles. Many studies have shown expression divergence between WGD-duplicates (17–22). However, a major obstacle encountered in all these studies is that they could not differentiate between sub and neofunctionalization because the pattern of expression before duplication was unknown. This obstacle was overcome recently for gene pairs that were formed by WGD in Saccharomyces cerevisiae by comparing their pattern of expression to Candida albicans, an outgroup whose genome was not duplicated and therefore can be used to approximate the ancestral expression state (23).
Here, we apply a similar approach to search for evidence of gene subfunctionalization after WGD in a vertebrate system. We compare the expression profiles of gene pairs preserved in duplicate after WGD in X. laevis to the expression profiles of orthologous genes in the unduplicated clawed frog S. tropicalis (sometimes also called X. tropicalis). The WGD that has been proposed for X. laevis has not yet been validated by a complete genome sequence, but it is estimated to have occurred 21–54.6 Mya (6, 24, 25) and it is likely to have been an allopolyploidization because interspecies crosses in Xenopus often produce fertile polyploid offspring and phylogenetic studies have shown that other polyploid clawed frogs are ancient allopolyploids (24, 26, 27).
We used the extensive expressed sequence tag (EST) and cDNA sequence resources available for these species (20, 21, 25) to detect genes present in one copy in S. tropicalis and in two copies in X. laevis. We inferred the pattern of expression in these triplets and detected events of subfunctionalization. We then tested whether the subfunctionalized genes are a random subset of the genome.
Results
Construction of the Dataset.
We clustered Xenopus expressed sequences (ESTs and full-length cDNAs) that are publicly available (558,503 sequences for X. laevis and 1,046,555 for S. tropicalis). We chose very stringent clustering parameters to avoid merging sequences expressed by paralogous genes [see Methods, supporting information (SI) Methods, and Fig. S1]. Using phylogenetic analysis, we built a dataset of 1,300 triplets, composed of one gene in S. tropicalis and its two coorthologs in X. laevis, whose duplication was most probably due to WGD.
An early study based on a very small dataset proposed that 77% of genes were retained in duplicate in X. laevis (28). By using highly expressed genes to minimize errors associated with EST sampling, we estimate that ≈32–47% of genes were retained in double-copy in X. laevis after WGD (SI Methods and Figs. S2 and S3). Our figure is similar to Hellsten et al.'s (21) estimate of ≈25–50% retention. Gene loss has been less extensive after the relatively recent WGD in X. laevis than after the teleost-specific WGD, which is 10 times older (29, 30): In Tetraodon nigroviridis for instance, only 15% of genes were retained in duplicate (7).
We estimated the gene expression profiles of each triplet based on the tissue from which the ESTs were extracted. More precisely, we obtained for each gene in each triplet a measure of its level of expression in each of 11 tissues that had been used for library construction in both species (see Methods for a list). We measured the conservation of these expression patterns between the two X. laevis copies since WGD by a Spearman correlation coefficient. We find that the majority of duplicate pairs do not show much divergence in expression since WGD (median correlation rho = 0.64; Fig. S4), a result similar to that of Chain et al. (22).
Detection of Changes in Expression Profile: Subfunctionalization and Asymmetric Changes.
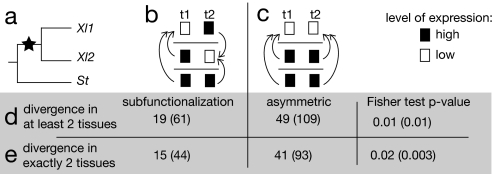
We used parsimony to estimate the pattern of evolution of expression in each triplet. The principle of our analysis is shown in Fig. 1. We say that a pair of duplicates in X. laevis has become subfunctionalized if we infer that one gene copy shows a significant decrease in expression level in one tissue, whereas the other copy shows a significant decrease in a different tissue (Fig. 1b). We modified slightly a statistical test developed by Audic and Claverie (31) to take into account the effect of WGD and subsequent gene losses on the relative contribution of each gene to the total pool of mRNA in the cell (see Methods, SI Methods, and Figs. S5 and S6). We performed this test on the 1,300 triplets and found 61 examples of subfunctionalization (4%). This number drops to 19 triplets (1.2%) if we correct for multiple testing [false discovery rate (FDR) < 0.05] (32). These triplets are loci in which expression has been significantly decreased in one X. laevis copy in one tissue and in the other X. laevis copy in another tissue, whatever other changes happened (significant or not) in the nine remaining tissues. Among these 19 triplets, 15 have undergone significant changes in exactly two tissues and no significant changes in the other tissues (44 without correction for multiple testing).
Fig. 1.
Principles of expression evolution in Xenopus sequence triplets. (a) Each triplet includes one gene in S. tropicalis (St) and its two coorthologs in X. laevis (Xl1 and Xl2) created by WGD. (b) A case of subfunctionalization. Columns t1 and t2 represent two tissues. Arrows represent the results of statistical tests and point to the gene with the significantly lower expression level. Here, expression of gene Xl1 in tissue t1 is significantly lower than expression of both St and Xl2 in the same tissue. In tissue t2, gene Xl2 shows lower expression. We infer that the gene was expressed in both tissues t1 and t2 before WGD and that subsequently the expression of Xl1 decreased in t1, whereas expression of its paralog Xl2 decreased in t2; this corresponds to a subfunctionalization pattern. (c) A case of asymmetric evolution of expression. Significant decreases in expression are inferred in X. laevis in two tissues, but they both involve the same X. laevis gene (here, Xl1). (d) Numbers of gene triplets showing subfunctionalization and asymmetric patterns of expression evolution as defined above. Numbers in parentheses do not include correction for multiple testing. P values by Fisher's test show that asymmetric evolution of expression is more frequent than subfunctionalization. (e) Numbers of triplets with subfunctionalized and asymmetric patterns of expression, defined as cases where there is a significant decrease of expression in exactly two tissues, and no significant decrease in the other nine tissues.
We implemented another method to identify subfunctionalization between the two X. laevis copies. We constructed for each triplet the pattern Xlsum by merging the patterns of expression of the two X. laevis copies (summing the number of ESTs per million for each tissue). Subfunctionalizations are cases where each of the copies in X. laevis has retained part of the ancestral function; therefore, the Spearman correlation of the patterns of expression between S. tropicalis and Xlsum should be higher than both of the correlations between S. tropicalis and the individual X. laevis genes. This pattern was found in 11% of the triplets (144 triplets).
Cases of subfunctionalization, therefore, represent only a small proportion (1.2–11%) of the WGD-duplicates considered here; however, we have seen that most pairs have not diverged in expression since the WGD (Fig. S4). We tested whether, among the minority of genes that do show significant changes in expression in our dataset, the pattern of changes frequently corresponds to a subfunctionalization pattern. We searched in particular for two patterns of expression profile change, which we refer to as subfunctionalization and asymmetric change (Fig. 1 b and c). Both of these patterns involve decreases of expression in X. laevis compared with S. tropicalis. We detected 49 cases of asymmetric partitioning of expression (109 without multiple testing), defined as triplets where expression has decreased significantly in at least two tissues since WGD, in the same X. laevis copy (Fig. 1c). Therefore, we estimate that, among X. laevis gene pairs whose expression has diverged significantly, one-third (19 of 68) are subfunctionalized, which is less than the 50% expected by chance (P = 0.01 by Fisher's test). This ratio remains constant if we only consider genes with significant changes in exactly two tissues (Fig. 1e).
Relationship Between Rate of Sequence Evolution and Pattern of Expression Divergence.
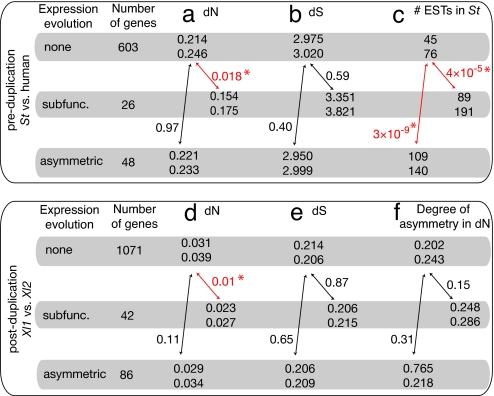
We examined whether the rate of evolution of a gene influences the evolution of its expression patterns after WGD. Because duplication tends to increase the rate of nonsynonymous sequence evolution [for instance, in Xenopus (21)], we instead measured this rate (dN) between two species whose genomes have not been duplicated: S. tropicalis and human. This dN value should be indicative of the gene's evolutionary rate before WGD. We find that genes that became subfunctionalized were more slowly evolving before WGD than the genes with no particular pattern of expression evolution (median dN values 0.154 and 0.214 respectively; P = 0.018 with two repetitions is significant at a 3.6% level; Fig. 2a). We consider this difference as biologically meaningful, even though its significance is marginal after Bonferroni correction, because the median dN is 40% lower in subfunctionalized genes than in the other genes, and because the power of the test is not very high given the small size of the datasets. In contrast, there is no significant rate difference between genes that later underwent an asymmetric pattern of expression evolution in X. laevis and those with no particular pattern of expression change (P = 0.97; Fig. 2a).
Fig. 2.
Rates of sequence evolution are correlated with expression evolution. Patterns of expression evolution are classified as subfunctionalized or asymmetric as in Fig. 1e; “none” refers to triplets that show neither of these patterns. dN, dS, and #ESTs are, respectively, the levels of nonsynonymous and synonymous sequence evolution and the number of ESTs in S. tropicalis (St). The median and mean of these variables are indicated (above and below, respectively), and P values for Wilcoxon tests for the pairwise comparison between the set “none” and each of the other sets of genes are shown near the arrows. Significant P values after Bonferroni correction (two test repetitions per panel) are indicated in red and marked with an asterisk. (a–c) Preduplication rates of sequence evolution are estimated between S. tropicalis and human. (d–f) Postduplication rates are computed between the two copies in X. laevis (Xl1 and Xl2).
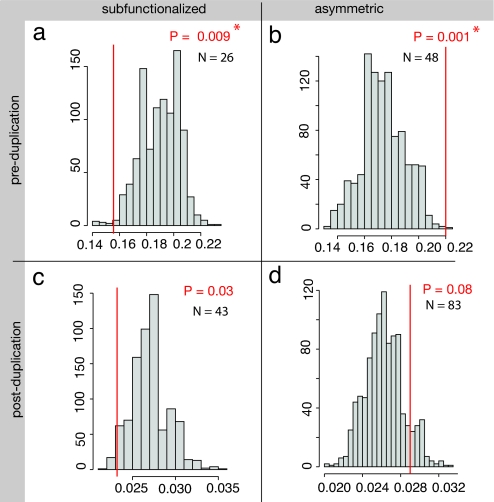
This preferential subfunctionalization of slowly evolving genes was unexpected. It is not a bias because of differences in mutation rate or in the age of the duplicates, because the levels of synonymous substitution are not significantly different among the three categories of genes (Fig. 2b). There is, however, another possible bias. By construction, the triplets with either subfunctionalization or asymmetric change of expression are expressed in more tissues and at a higher level than triplets that do not show these patterns (Fig. 2c), and it is known that genes expressed in many tissues evolve more slowly than other genes (33, 34). However, after correcting for this bias we still find that genes that became subfunctionalized are descended from exceptionally slowly evolving ancestors (Fig. 3a; P = 0.009 with four repetitions is significant at a 3.6% level). Moreover, this method of analysis also shows an opposite pattern in genes that developed asymmetric expression patterns after WGD: The ancestors of these genes were unusually quickly evolving (Fig. 3b; P = 0.001 with four repetitions is significant at a 0.4% level). These results are very surprising because they show that the rate of sequence evolution before the duplication influences the pattern of evolution of expression after the duplication.
Fig. 3.
The rate of nonsynonymous sequence evolution (dN) before the duplication influences the pattern of evolution of expression after the duplication. Shown are comparisons of an observed median dN value (red line) with a histogram of the distribution of expected values; the x axis in each panel is in dN units, and the y axis shows the number of genes in the histogram. P values for the comparisons between the observed values and the distributions are shown. Asterisks mark tests that are significant at a 5% level after Bonferroni correction for the four test repetitions. (a) The median value (0.154) of dN(human, S. tropicalis) for the set of 26 genes that show subfunctionalization in X. laevis and have an annotated ortholog in human is superimposed on a histogram showing the distribution of median values of dN(human, S. tropicalis) obtained from 1,000 samples. Each of these samples contains 100 genes chosen randomly from triplets with neither subfunctionalized nor asymmetric pattern of evolution of expression, such that the distribution of the levels of expression (measured in S. tropicalis) in each sample is the same as in the distribution observed in the set of 26 subfunctionalized genes. (b–d) Comparisons of observed median dN values with histograms of the distributions of median dN values obtained from same-size samples of genes showing no significant expression evolution and after correction for expression level. The four panels compare the observed median dN values (red line) of loci whose X. laevis coorthologs show either subfunctionalization (a and c) or asymmetric (b and d) patterns of expression evolution, to distributions sampled from triplets that do not show such patterns and after correction for expression level. a and b show dN values calculated between S. tropicalis and human, representing the preduplication rate of evolution, and c and d show dN values calculated from comparisons between the X. laevis coorthologs, representing the postduplication rate.
We then asked whether the triplets of genes with subfunctionalization or asymmetric patterns of expression evolution have a particular rate of evolution after WGD. We computed the nonsynonymous divergence between the two paralogous copies in X. laevis and compared the mean values among groups with different patterns of expression evolution (Fig. 2d). Nonsynonymous divergence between X. laevis copies is significantly smaller for subfunctionalized genes (median dN: 0.023) than for genes with no particular pattern of expression evolution (median dN: 0.031; P = 0.01 by Wilcoxon test with two repetitions is significant at a 2% level). This effect is only marginally significant after correction for expression bias (Fig. 3c, P = 0.03 with four repetitions is marginally significant at a 12% level). Genes with an asymmetric pattern of evolution do not have a particular rate of evolution after duplication (Fig. 3d; P = 0.08 is not significant after Bonferroni correction). We also evaluated the asymmetry in the rates of nonsynonymous substitution between the two copies in X. laevis, using like-tri-test (35), but we found no link between sequence asymmetry and the pattern of expression evolution (Fig. 2f). A recent study showed that rates of nonsynonymous sequence evolution increased after WGD in X. laevis sequences, but this was a small effect only visible after concatenation of the sequences (21). Therefore, it is possible that nonsynonymous rates of evolution are asymmetric after duplication, but this effect is not visible on a gene-by-gene basis.
Convergent Outcomes of Two Independent WGDs in Teleost Fish and X. laevis.
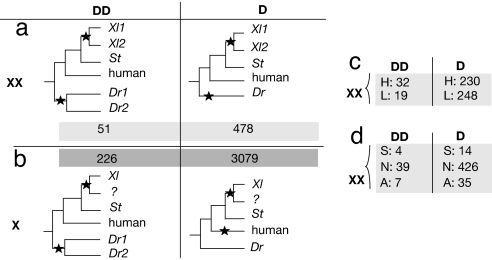
We have seen that subfunctionalized genes in X. laevis are distinctive because they were slowly evolving before WGD. Therefore, it is possible that some genes are more prone to subfunctionalization than others. To test this hypothesis, we compared the outcomes of two WGDs that occurred independently in vertebrates: one in X. laevis and one at the base of teleost fish lineage. First, we tested the null hypothesis that the two WGDs should have independent results in terms of double-copy retentions. In other words, whether a gene pair was or was not retained in duplicate in X. laevis should have no bearing on whether or not its orthologous pair was retained in fish. Because the genome of zebrafish has been completely sequenced, we can assess with certainty whether a gene pair was retained in two copies in this species after its WGD, whereas this is not feasible in X. laevis. We identified reliable orthologs in zebrafish for half of our triplets (529 genes; Fig. 4a). The fraction of these families that contain two WGD coorthologs in zebrafish is 9.5% (51 of 529). This fraction is significantly higher than the corresponding fraction for gene families in which only one X. laevis copy was found (6.8%; 226 of 3,305; Fig. 4b; P = 0.036 by Fisher's test). The latter set corresponds to families for which we did not detect a second gene in X. laevis, which in some cases might have been because no EST was sampled rather than because the gene is truly single-copy in X. laevis. For this reason, our test is a conservative one, and we conclude that genes retained in duplicate after one vertebrate WGD event have increased probability of also having been retained after the other.
Fig. 4.
Comparison of genes retained in double-copy in X. laevis and in zebrafish shows that the two independent WGDs do not have independent outcomes. (a) 529 triplets with two copies in X. laevis (designated XX) were sorted into groups whose orthologs in zebrafish are double-copy or single-copy (designated DD and D, respectively). Zebrafish gene pairs (Dr1 and Dr2) in the same Homolens family were attributed to the teleost WGD if their duplication is older than the speciation of zebrafish and pufferfish. (b) 3305 doublets (designated X) for which we could not find a second copy in X. laevis were sorted similarly according to their duplication status in zebrafish. (c) Distribution of the 529 XX triplets by their expression level in S. tropicalis (two classes: H, high; L, low) and their duplication status in zebrafish. (d) Distribution of the 529 XX triplets by their pattern of expression evolution (three classes: S, subfunctionalization; A, asymmetric partitioning; N, neither) and their duplication status in zebrafish.
What is the reason for this convergence? It has been shown that highly expressed genes are overretained after WGD (15, 36). If expression level differences are responsible for the nonindependence of the two WGDs, we would expect that highly expressed genes should have higher frequencies of retention in duplicate than weakly expressed genes, after both WGDs. We divided our dataset of Xenopus-fish orthologs into two classes depending on their expression level in S. tropicalis (low or high; Fig. 4c) and observed that the proportion of genes retained in duplicate in zebrafish is higher (12%; 32 of 262) for genes highly expressed in S. tropicalis than for low-expression genes (7%; 19 of 267; P = 0.04 by Fisher's test). The genes responsible for the nonindependence of the two duplications are therefore highly expressed.
We can ask whether these highly expressed genes have been retained for the same reason after the two WGDs. By definition, subfunctionalized genes are expressed in several tissues and they are also highly expressed (Fig. 2c), so it is possible that the genes convergently retained in duplicate after the two WGDs are enriched in subfunctionalized genes. Indeed, we find that genes that have been subfunctionalized in X. laevis have a higher frequency of parallel retention in zebrafish (22%; 4 of 18; Fig. 4d) than do those that show no pattern of expression divergence or an asymmetric divergence pattern (8 and 16%, respectively; data from Fig. 4d; P = 0.037 by χ2 test of homogeneity among these three categories). These results suggest that gene pairs retained by subfunctionalization in X. laevis also tended to be retained by subfunctionalization in zebrafish. Unfortunately, we cannot directly test whether these pairs have been subfunctionalized in zebrafish, because no expression data are available for any outgroup species that diverged shortly before the teleost WGD. However, our hypothesis of convergent subfunctionalization receives some support from a comparison of the divergence of expression profiles between the pairs in zebrafish and in X. laevis (Fig. S7).
Discussion
In their pioneering study of 17 duplicated genes in X. laevis, Hughes and Hughes (28) already noticed that in four cases the two copies were expressed in different tissues or at different developmental times, and this trend was confirmed recently in larger datasets (20, 21). We detect relatively little subfunctionalization in our dataset (1.2–11% of the WGD-duplicates considered). This may be because most pairs have not diverged in expression since the WGD and subfunctionalization actually only happened in a small percentage of pairs. Alternatively, our ability to detect subfunctionalization is perhaps limited. If we had complete information about transcription in every tissue, we could more accurately detect significant expression divergence between gene pairs in some particular tissues and hence obtain a reliable estimate of the fraction of genes undergoing subfunctionalization. The frequency of subfunctionalization we estimate here is a lower limit, because we examined a limited number of tissues, but we cannot propose an upper bound for this figure. Even though we were not able to estimate the frequency of subfunctionalization, we could still examine the characteristics of genes that became subfunctionalized.
We find that some genes are predisposed to subfunctionalization. Genes that underwent subfunctionalization in X. laevis tend to be slowly evolving in other species, and conversely genes with an asymmetric pattern of expression evolution in X. laevis tend to evolve faster than expected in these outgroups. These results are only of medium statistical significance, partly because the limited size of the datasets weakens the power of the tests, and partly because of the necessity to correct for multiple testing. Nonetheless, we observed that the rate of sequence evolution influences the retention of some genes after WGD, and we propose a model to explain our observations.
Genes retained in duplicate after WGD are more likely to belong to gene families with slow rates of sequence evolution (7, 37, 38) or high expression levels (15, 36). Slow sequence evolution is correlated with a high level (or wide breadth) of expression in both yeast and vertebrates (33, 39), and both observations may be due to the same phenomenon. Davis and Petrov (37) were unable to find an obvious explanation for their discovery that slowly evolving genes are preferentially retained in duplicate, but they suggested that the bias may be an indirect correlation due to a third variable that is responsible for the retention and is correlated with the other two. Candidates for this third variable include the presence of many cis-regulatory regions (5, 40), of genes coding for multidomain proteins (41), and pleiotropic genes (model 3 in ref. 42). Other models predict that genes with a particular function, such as regulatory genes (43), should be retained in duplicate more often than expected after WGD. Alternatively there may be a direct relationship between the expression level (or the rate of sequence evolution) and the propensity to be retained in duplicate. Highly expressed genes may be retained in duplicate after WGD simply because they are beneficial for gene dosage (15). We discuss below that the rate of evolution seems to be directly responsible for double-copy retention in Xenopus, at least for the subset of gene pairs whose expression is divergent.
We have shown that slowly evolving genes are more subject to subfunctionalization. Theoretical studies of subfunctionalization do not predict that genes becoming subfunctionalized should evolve more slowly than others before duplication (5, 40). On the contrary, subfunctionalization is supposed to be a neutral event that occurs because of neutral mutations impairing different subfunctions in the duplicates. Because we correct for expression bias (Fig. 3), we show more exactly that among a pool of genes with the same level of expression, slowly evolving genes are more likely to be subfunctionalized, and fast evolving genes are more likely to have an asymmetric pattern of expression evolution. We can extrapolate that genes that are subfunctionalized are going to be retained in two copies in the genome (both copies are necessary to perform the ancestral function), but, in contrast, it is likely that many genes with an asymmetric pattern of evolution of expression will eventually return to single-copy state. This is confirmed by our comparison between WGDs in Xenopus and zebrafish: subfunctionalized genes in Xenopus, but not genes with an asymmetric pattern of expression evolution, are retained in two copies more than expected in zebrafish.
These observations lead us to propose that slowly evolving genes were more easily subfunctionalized in X. laevis and therefore more easily retained long after WGD. Our model of gene evolution after WGD in X. laevis is illustrated in Fig. 5, which is based on the assumption that the WGD was an allopolyploidization, as is most likely (26, 27). Most models to explain gene retention after WGD postulate that the two copies are equal at birth (e.g., refs. 5, 43) but this is not true in the case of allopolyploidization. In our model, two diverging populations accumulate sequence differences, but to a greater extent in faster-evolving genes than in slower-evolving genes. When their two genomes are merged by allopolyploidization, the slower-evolving loci have accumulated fewer substitutions so the two copies may still be interchangeable and subfunctionalization can occur. In contrast, in the faster-evolving genes it is more likely that there are functional differences between the two copies, and one of them functions better than the other. If so, it may be deleterious for the better-functioning gene copy to lose any of its subfunctions. Such a situation will prevent subfunctionalization from happening; instead, the worse-functioning gene copy will be lost completely. In X. laevis, we observe the consequences of an allopolyploidization of medium age, where nearly half the genes are still in duplicate. For the fast-evolving genes in this genome we tend to see an asymmetry in the expression patterns and we anticipate that the worse-functioning copy will be lost eventually. Our observations and our model contradict a previous hypothesis by Spring (44), who suggested that slower-evolving genes would be more redundant at the time of allopolyploidization and therefore easier to lose.
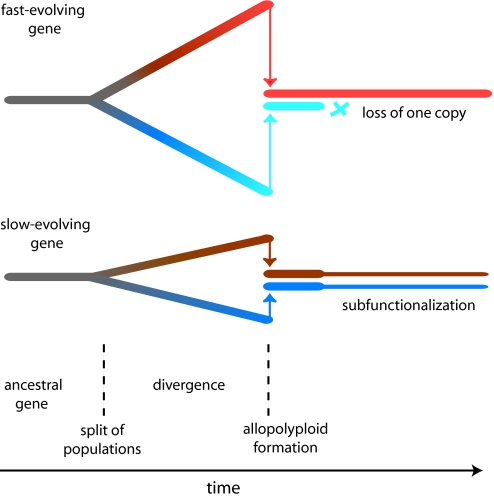
Fig. 5.
Model to explain why slowly evolving genes are preferentially subfunctionalized after allopolyploidization. The horizontal axis represents time. Initially two populations diverge and their genes begin to accumulate sequence differences, represented by color changes and vertical separation. They subsequently hybridize to form an allopolyploid. For a fast-evolving gene (Upper), the two copies are very different at the time of allopolyploid formation. They are unlikely to be functionally indistinguishable, and it is probable that only the better-functioning one of them will be retained. For a slow-evolving gene (Lower), the two copies are less different and more likely to be able to replace one another functionally. Subfunctionalization (represented by thinning of the lines) may result.
Note that our model is also valid for an autopolyploidization or any other kind of gene duplication, if the genes can survive in duplicate long enough to attain sequence divergence. In each case the genes with slower rates of nonsynonymous substitution are expected to remain equivalent (and therefore prone to subfunctionalization) for a longer time. This hypothesis is supported by our laboratory's previous work on WGD in yeast, where we found that slow-evolving genes retained their interchangeability for a longer time period after WGD than fast-evolving genes (45, 46). If this idea is correct it can account for the preferential retention of slow-evolving genes after any kind of duplication, as seen by Davis and Petrov (37). Thus, subfunctionalization may be a force in the long-term evolution of duplicated genes, in addition to its originally postulated role (5) in their initial preservation.
Methods
The methods we used for stringent EST clustering, building triplets of homologous Xenopus genes, establishing orthology relationships, and estimating rates of sequence evolution are described in SI Methods. The derivation of our estimate that 32–47% of genes were retained in double-copy in X. laevis after WGD is also given in SI Methods.
To estimate expression profiles of frog genes, we classified the available Xenopus EST libraries into tissues (104 libraries in X. laevis, 51 in S. tropicalis) and identified the following 11 tissues (or developmental stages) as being common between the two species: brain, embryo, heart, kidney, liver, lung, ovary, skin, spleen, tadpole, and testis. By construction, each contig in a triplet is composed of ESTs that were used to infer its pattern of expression. Zebrafish EST analysis is described in SI Methods.
To detect differences in expression level between the two copies in X. laevis (denoted Xl1 and Xl2) and S. tropicalis (St) in one tissue, we used Audic and Claverie's Bayesian test (31), which takes the total number of ESTs sequenced in each tissue from each species into account. We modified the test slightly because the null expectation is that the EST count of gene St should be ≈1.3 times greater (exact value: e0.26; see SI Methods) than the individual EST counts of its orthologs Xl1 and Xl2. To detect a significant decrease in the expression of gene Xl1 in a particular tissue we tested whether, for this tissue in the two species, (i) the EST count of Xl1 × e0.26 is significantly lower than the count of St, and (ii) the EST count of Xl2 × e0.26 is not significantly lower than the count of St.
Supplementary Material
Acknowledgments.
We thank G. Conant for help with computing of asymmetry of sequence evolution; L. Guéguen for help with FDR and tree parsing; and K. Byrne, G. Conant, B. Cusack, C. Frank, J. Gordon, N. Khaldi, J. Mower, D. Scannell, M. Webster, M. Woolfit, and two anonymous referees for helpful comments. This work was supported by Irish Research Council for Science, Engineering and Technology and the Science Foundation Ireland.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708705105/DCSupplemental.
References
- 1.Otto SP, Whitton J. Polyploid incidence and evolution. Annu Rev Genet. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- 2.Jaillon O, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- 3.Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature. 2006;440:341–345. doi: 10.1038/nature04562. [DOI] [PubMed] [Google Scholar]
- 4.Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chain FJ, Evans BJ. Multiple mechanisms promote the retained expression of gene duplicates in the tetraploid frog Xenopus laevis. PLoS Genet. 2006;2:e56. doi: 10.1371/journal.pgen.0020056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunet FG, et al. Gene loss and evolutionary rates following whole-genome duplication in teleost fishes. Mol Biol Evol. 2006;23:1808–1816. doi: 10.1093/molbev/msl049. [DOI] [PubMed] [Google Scholar]
- 8.Steinke D, Salzburger W, Braasch I, Meyer A. Many genes in fish have species-specific asymmetric rates of molecular evolution. BMC Genomics. 2006;7:20. doi: 10.1186/1471-2164-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altschmied J, et al. Subfunctionalization of duplicate mitf genes associated with differential degeneration of alternative exons in fish. Genetics. 2002;161:259–267. doi: 10.1093/genetics/161.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cresko WA, et al. Genome duplication, subfunction partitioning, and lineage divergence: Sox9 in stickleback and zebrafish. Dev Dyn. 2003;228:480–489. doi: 10.1002/dvdy.10424. [DOI] [PubMed] [Google Scholar]
- 11.Yu WP, Brenner S, Venkatesh B. Duplication, degeneration and subfunctionalization of the nested synapsin-Timp genes in Fugu. Trends Genet. 2003;19:180–183. doi: 10.1016/S0168-9525(03)00048-9. [DOI] [PubMed] [Google Scholar]
- 12.de Souza FS, Bumaschny VF, Low MJ, Rubinstein M. Subfunctionalization of expression and peptide domains following the ancient duplication of the proopiomelanocortin gene in teleost fishes. Mol Biol Evol. 2005;22:2417–2427. doi: 10.1093/molbev/msi236. [DOI] [PubMed] [Google Scholar]
- 13.Chang L, Khoo B, Wong L, Tropepe V. Genomic sequence and spatiotemporal expression comparison of zebrafish mbx1 and its paralog, mbx2. Dev Genes Evol. 2006;216:647–654. doi: 10.1007/s00427-006-0082-7. [DOI] [PubMed] [Google Scholar]
- 14.Cusack BP, Wolfe KH. When gene marriages don't work out: Divorce by subfunctionalization. Trends Genet. 2007;23:270–272. doi: 10.1016/j.tig.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Aury JM, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- 16.Woolfe A, Elgar G. Comparative genomics using Fugu reveals insights into regulatory subfunctionalization. Genome Biol. 2007;8:R53. doi: 10.1186/gb-2007-8-4-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casneuf T, De Bodt S, Raes J, Maere S, Van de Peer Y. Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana. Genome Biol. 2006;7:R13. doi: 10.1186/gb-2006-7-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16:1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duarte JM, et al. Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol Biol Evol. 2006;23:469–478. doi: 10.1093/molbev/msj051. [DOI] [PubMed] [Google Scholar]
- 20.Morin RD, et al. Sequencing and analysis of 10,967 full-length cDNA clones from Xenopus laevis and Xenopus tropicalis reveals post-tetraploidization transcriptome remodeling. Genome Res. 2006;16:796–803. doi: 10.1101/gr.4871006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hellsten U, et al. Accelerated gene evolution and subfunctionalization in the pseudotetraploid frog Xenopus laevis. BMC Biol. 2007;5:31. doi: 10.1186/1741-7007-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chain FJ, Ilieva D, Evans BJ. Duplicate gene evolution and expression in the wake of vertebrate allopolyploidization. BMC Evol Biol. 2008;8:43. doi: 10.1186/1471-2148-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tirosh I, Barkai N. Comparative analysis indicates regulatory neofunctionalization of yeast duplicates. Genome Biol. 2007;8:R50. doi: 10.1186/gb-2007-8-4-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evans BJ, Kelley DB, Melnick DJ, Cannatella DC. Evolution of RAG-1 in polyploid clawed frogs. Mol Biol Evol. 2005;22:1193–1207. doi: 10.1093/molbev/msi104. [DOI] [PubMed] [Google Scholar]
- 25.Pollet N, Mazabraud A. In: Genome Dynamics, Volume 2, Vertebrate Genomes. Volff J-N, editor. Basel, Switzerland: Karger; 2006. pp. 138–153. [DOI] [PubMed] [Google Scholar]
- 26.Kobel HR. In: The Biology of Xenopus. Tinsley RC, Kobel HR, editors. Oxford: Clarendon; 1996. pp. 391–401. [Google Scholar]
- 27.Evans BJ. Ancestry influences the fate of duplicated genes millions of years after polyploidization of clawed frogs (Xenopus) Genetics. 2007;176:1119–1130. doi: 10.1534/genetics.106.069690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes MK, Hughes AL. Evolution of duplicate genes in a tetraploid animal, Xenopus laevis. Mol Biol Evol. 1993;10:1360–1369. doi: 10.1093/oxfordjournals.molbev.a040080. [DOI] [PubMed] [Google Scholar]
- 29.Vandepoele K, De Vos W, Taylor JS, Meyer A, Van de Peer Y. Major events in the genome evolution of vertebrates: Paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc Natl Acad Sci USA. 2004;101:1638–1643. doi: 10.1073/pnas.0307968100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoegg S, Brinkmann H, Taylor JS, Meyer A. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J Mol Evol. 2004;59:190–203. doi: 10.1007/s00239-004-2613-z. [DOI] [PubMed] [Google Scholar]
- 31.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 32.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A Practical and powerful approach to multiple testing. J Roy Stat Soc B. 1995;57:289–300. [Google Scholar]
- 33.Duret L, Mouchiroud D. Determinants of substitution rates in mammalian genes: Expression pattern affects selection intensity but not mutation rate. Mol Biol Evol. 2000;17:68–74. doi: 10.1093/oxfordjournals.molbev.a026239. [DOI] [PubMed] [Google Scholar]
- 34.Zhang L, Li WH. Mammalian housekeeping genes evolve more slowly than tissue-specific genes. Mol Biol Evol. 2004;21:236–239. doi: 10.1093/molbev/msh010. [DOI] [PubMed] [Google Scholar]
- 35.Conant GC, Wagner A. Asymmetric sequence divergence of duplicate genes. Genome Res. 2003;13:2052–2058. doi: 10.1101/gr.1252603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seoighe C, Wolfe KH. Yeast genome evolution in the post-genome era. Curr Opin Microbiol. 1999;2:548–554. doi: 10.1016/s1369-5274(99)00015-6. [DOI] [PubMed] [Google Scholar]
- 37.Davis JC, Petrov DA. Preferential duplication of conserved proteins in eukaryotic genomes. PLoS Biol. 2004;2:E55. doi: 10.1371/journal.pbio.0020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis JC, Petrov DA. Do disparate mechanisms of duplication add similar genes to the genome? Trends Genet. 2005;21:548–551. doi: 10.1016/j.tig.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 39.Drummond DA, Raval A, Wilke CO. A single determinant dominates the rate of yeast protein evolution. Mol Biol Evol. 2006;23:327–337. doi: 10.1093/molbev/msj038. [DOI] [PubMed] [Google Scholar]
- 40.Lynch M, O'Hely M, Walsh B, Force A. The probability of preservation of a newly arisen gene duplicate. Genetics. 2001;159:1789–1804. doi: 10.1093/genetics/159.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gibson TJ, Spring J. Genetic redundancy in vertebrates: Polyploidy and persistence of genes encoding multidomain proteins. Trends Genet. 1998;14:46–49. doi: 10.1016/s0168-9525(97)01367-x. [DOI] [PubMed] [Google Scholar]
- 42.Nowak MA, Boerlijst MC, Cooke J, Smith JM. Evolution of genetic redundancy. Nature. 1997;388:167–171. doi: 10.1038/40618. [DOI] [PubMed] [Google Scholar]
- 43.Birchler JA, Pal-Bhadra M, Bhadra U. Dosage dependent gene regulation and the compensation of the X chromosome in Drosophila males. Genetica. 2003;117:179–190. doi: 10.1023/a:1022935927763. [DOI] [PubMed] [Google Scholar]
- 44.Spring J. Vertebrate evolution by interspecific hybridisation—are we polyploid? FEBS Lett. 1997;400:2–8. doi: 10.1016/s0014-5793(96)01351-8. [DOI] [PubMed] [Google Scholar]
- 45.Scannell DR, et al. Independent sorting-out of thousands of duplicated gene pairs in two yeast species descended from a whole-genome duplication. Proc Natl Acad Sci USA. 2007;104:8397–8402. doi: 10.1073/pnas.0608218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Byrne KP, Wolfe KH. Consistent patterns of rate asymmetry and gene loss indicate widespread neofunctionalization of yeast genes after whole-genome duplication. Genetics. 2007;175:1341–1350. doi: 10.1534/genetics.106.066951. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.