Abstract
Since the first use of streptomycin as an effective antibiotic drug in the treatment of tuberculosis, aminoglycoside antibiotics have been widely used against a variety of bacterial infections for over six decades. However, the pathways for aminoglycoside biosynthesis still remain unclear, mainly because of difficulty in genetic manipulation of actinomycetes producing this class of antibiotics. Gentamicin belongs to the group of 4,6-disubstituted aminoglycosides containing a characteristic core aminocyclitol moiety, 2-deoxystreptamine (2-DOS), and the recent discovery of its biosynthetic gene cluster in Micromonospora echinospora has enabled us to decipher its biosynthetic pathway. To determine the minimal set of genes and their functions for the generation of gentamicin A2, the first pseudotrisaccharide intermediate in the biosynthetic pathway for the gentamicin complex, various sets of candidate genes from M. echinospora and other related aminoglycoside-producing strains were introduced into a nonaminoglycoside producing strain of Streptomyces venezuelae. Heterologous expression of different combinations of putative 2-DOS biosynthetic genes revealed that a subset, gtmB-gtmA-gacH, is responsible for the biosynthesis of this core aminocyclitol moiety of gentamicin. Expression of gtmG together with gtmB-gtmA-gacH led to production of 2′-N-acetylparomamine, demonstrating that GtmG acts as a glycosyltransferase that adds N-acetyl-d-glucosamine (GLcNA) to 2-DOS. Expression of gtmM in a 2′-N-acetylparomamine-producing recombinant S. venezuelae strain generated paromamine. Expression of gtmE in an engineered paromamine-producing strain of S. venezuelae successfully generated gentamicin A2, indicating that GtmE is another glycosyltransferase that attaches d-xylose to paromamine. These results represent in vivo evidence elucidating the complete biosynthetic pathway of the pseudotrisaccharide aminoglycoside.
Keywords: aminoglycoside biosynthesis, Micromonospora echinospora, Streptomyces venezuelae, pathway engineering
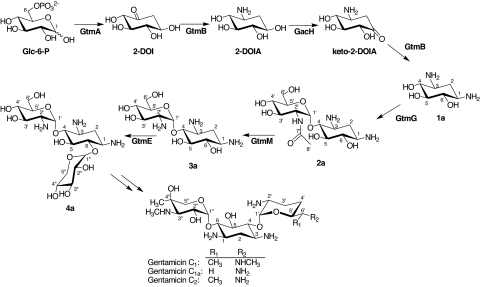
Gentamicin is an aminoglycoside antibiotic complex produced by various Micromonospora species (1). Aminoglycosides act by binding to the bacterial ribosome inhibiting protein synthesis and generating errors in the translation of the genetic code (2). This class of antibiotics has been used clinically for the treatment of severe bacterial infections for >60 years, beginning with the use of streptomycin to treat tuberculosis (3). However, the genetics underlying the biosynthesis of aminoglycosides, including gentamicin, still remains unclear. Structurally, gentamicin is a 4,6-disubstituted aminocyclitol composed of the core aminocyclitol moiety, 2-deoxystreptamine (2-DOS, 1a), which is decorated by purpurosamine and garosamine aminosugars at positions C-4 and C-6, respectively. In its therapeutic form, gentamicin comprises a complex of gentamicin C1, C1a, and C2, which differ only in the degree of methylation of the C-6′ position of the sugar attached at C-4 of 2-DOS (Fig. 1). Other minor components of gentamicin are also produced by Micromonospora echinospora, including gentamicin A2 (4a), the precursor from which other forms of gentamicin are derived (4).
Fig. 1.
Proposed biosynthetic pathway of gentamicin A2 (4a) and its biosynthetic intermediates 2-deoxystreptamine (1a), 2′-N-acetylparomamine (2a), and paromamine (3a).
The recent discovery of gene clusters for biosynthesis of butirosin and neomycin in Bacillus circulans and Streptomyces fradiae, respectively, has facilitated genetic and biochemical studies on the biosynthesis of 4,5-disubstituted aminoglycosides (5–9). Concurrently, the biosynthetic gene clusters relating to the 4,6-disubstituted aminoglycosides kanamycin, tobramycin, and gentamicin have been isolated from Streptomyces kanamyceticus, Streptomyces tenebrarius, and M. echinospora, respectively (10–13). However, little progress has been made to evaluate the proposed biosynthetic routes to these antibiotics, mainly because of difficulty in genetic manipulation of the producing microorganisms. Gentamicin-producing M. echinospora has innate resistance to most antibiotics precluding the use of standard Streptomyces cloning vectors displays low transformation efficiency (<10−7 transformants per microgram of DNA) (14). Current knowledge regarding the biosynthesis of 2-DOS containing aminoglycosides was recently reviewed, and most of cited works relied on in vitro studies of individual gene products (15). When the biosynthetic route to the gentamicin complex was first predicted, it was based on isolation of metabolites from cell culture extracts and precursor feeding studies of blocked mutants (16, 17). However, functional studies of gentamicin biosynthetic genes have not been possible, so that assignment of the roles of gene products has hitherto been based purely on amino acid sequence comparisons pending verification by direct experimental evidence.
We show here that heterologous expression of different combinations of genes from the gentamicin biosynthetic gene cluster (refs. 11 and 12: GenBank accession nos. AJ575934 and AY524043) in a nonaminoglycoside producing strain of S. venezuelae caused production of intermediates from the gentamicin biosynthetic pathway including 2-DOS (1a), 2′-N-acetylparomamine (2a), paromamine (3a), and the first pseudotrisaccharide, gentamicin A2 (4a). This provided experimental evidence for assigning the functions of individual gene products, allowing us to recognize a complete biosynthetic pathway for generation of gentamicin A2. This work has established an efficient heterologous expression platform based on a genetically amenable streptomycete to dissect the biosynthetic pathways of other aminoglycoside antibiotics produced by less tractable actinomycetes.
Results and Discussion
Putative gentamicin biosynthetic genes, suggested by previous studies to encode the 2-DOS biosynthetic enzymes, two glycosyltransferases and a deacetylase, were amplified by PCR (8, 10, 18) and cloned into a thiostrepton-resistance vector, pSE34, under control of the ermE* promoter (19). Plasmids harboring different sets of genes necessary for the biosynthesis of gentamicin and its intermediates were heterologously expressed in a mutant strain of S. venezuelae YJ003 that lacks the ability to biosynthesize its native deoxysugar (TDP-d-desosamine) (20). The culture extracts from the resulting strains were analyzed by using HPLC-ESI-MS/MS to detect gentamicin-related compounds, the structures of which were then further identified by comparison of the retention times, MS/MS fragmentation patterns, and NMR spectra with those obtained from authentic reference compounds.
A Minimal Set of Genes Necessary and Sufficient for 2-Deoxystreptamine Biosynthesis.
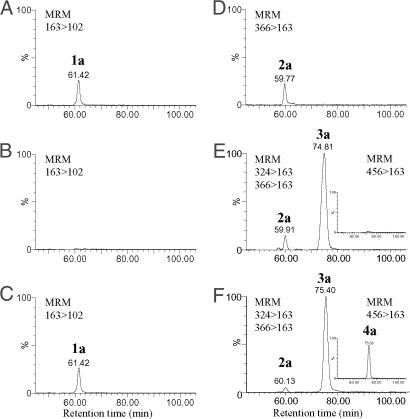
Gentamicin biosynthesis begins with d-glucose-6-phosphate (Glc-6-P), which is modified to 2-deoxy-scyllo-inosose (2-DOI) by carbocyclization through the action of GtmA, as described in ref. 12. Two aminotransferase reactions and a dehydrogenase step are then required for the conversion of 2-DOI via 2-deoxy-scyllo-inosamine (2-DOIA) and 1-keto-2-deoxy-scyllo-inosamine (keto-2-DOIA) to produce 2-DOS (1a) (21–24) (Fig. 1). Based on an earlier study isolating the gentamicin biosynthetic gene cluster (11, 12), we cloned the gtmD-gtmB-gtmA-gtmC genes, which encode the inosose synthase (GtmA) and putative aminotransferases (GtmB and GtmD), plus the deduced dehydrogenase (GtmC), and expressed them in the heterologous host S. venezuelae YJ003 using pYJ494 in what proved to be an unsuccessful attempt to biosynthesize 2-DOS (1a). This negative result prompted us to replace gtmC with gacH (otherwise known as gntP), which locates between gtmK and gacI within the gentamicin biosynthetic gene cluster, following the prediction (15) that GacH might be the dehydrogenase involved in 2-DOS biosynthesis, based on its sequence homology to the 2-DOIA dehydrogenase Neo5 [CAH58688, otherwise known as NeoA (BAD95818)] involved in neomycin biosynthesis. When the resultant plasmid pYJ495 harboring gtmD-gtmB-gtmA-gacH was expressed in the heterologous host, 2-DOS (1a) was indeed produced (Fig. 2A). The two transamination reactions required for biosynthesis of 2-DOS during butirosin production by B. circulans are catalyzed by a single enzyme, BtrS [BAE07061, otherwise known as BtrR (CAD41947)] (23). To ascertain whether the biosynthesis of 2-DOS during gentamicin production likewise involves dual activity of a single aminotransferase, two separate constructs pYJ496 and pYJ497 expressing gtmB-gtmA-gacH and gtmD-gtmA-gacH, respectively, were tested. The expression of gtmB, gtmA, and gacH produced ≈120 μg per liter of 2-DOS (1a) (corrected for ≈40% mean recoveries of 1a), whereas, gtmD, gtmA, and gacH did not do so (Fig. 2 B and C), indicating that GtmB catalyzes both transamination steps in the synthesis of 2-DOS, whereas GtmD may act in a later step during gentamicin production. The lower recoveries (<50%) of both 2-DOS (1a) and 2′-N-acetylparomamine (2a) were observed compared with those of other biosynthetic intermediates of gentamicin, probably because of their weak adsorption on the solid-phase extraction (SPE) cleanup column during analysis (25). The predicted products of gtmA, gtmB, and gacH are similar to those of other 2-DOS (1a) biosynthetic genes from several 4,5- and 4,6-disubstituted aminoglycoside producers [see supporting information (SI) Figs. S1–S3 and SI Text]. Interestingly, the deduced product of gacH, which contains two zinc-binding motifs found in the NAD(P)-dependent dehydrogenase NeoA (Neo5) (22), does not show sequence similarity to the recently characterized AdoMet-dependent dehydrogenase BtrN (24), suggesting that GacH belongs to the NAD(P)-dependent dehydrogenase family (see Fig. S3).
Fig. 2.
HPLC-ESI-MS/MS analysis of extracts from S. venezuelae strains harboring the plasmids (A) pYJ495 expressing gtmD-gtmB-gtmA-gacH. (B) pYJ497 expressing gtmD-gtmA-gacH. (C) pYJ496 expressing gtmB-gtmA-gacH. (D) pYJ498 expressing gtmB-gtmA-gacH-gtmG. (E) pYJ503 expressing gtmB-gtmA-gacH-gtmG-gtmM. (F) pYJ505 expressing gtmB-gtmA-gacH-gtmG-gtmE-gtmM. pYJ503 and pYJ505 coexpress the resistance genes gtmF-gtmL-gtmK. Monitoring of gentamicin A2 (4a) (456 > 163) is separately illustrated as an Inset to each chromatogram E and F.
Conferring Gentamicin Resistance in the Heterologous Host.
A prerequisite for the biosynthesis of gentamicin in a heterologous host is self-protection of the host strain. To confer resistance to gentamicin in S. venezuelae, a plasmid, pYJ489, harboring three putative resistance genes (gtmF-gtmL-gtmK), was introduced into this mutant strain. The resistance genes gtmF and gtmL were chosen based on their ability to confer gentamicin resistance via specific methylation of the bacterial ribosomes without modifying the antibiotic itself (11). The gtmK gene presumably encodes a transport protein involved in the export of aminoglycosides (11). Transformants harboring pYJ489 were resistant to 100 μg/ml of the commercially available gentamicin sulfate compared with ≈1 μg/ml in control strains. The cell culture was extracted and analyzed for gentamicin components, and any of their derivatives (but only unmodified gentamicins) were recovered. These resistance determinants were expressed together with the gentamicin biosynthetic genes in the heterologous host in subsequent experiments.
GtmG Acts as an UDP-GlcNAc Glycosyltransferase to Generate 2′-N-Acetylparomamine (2a).
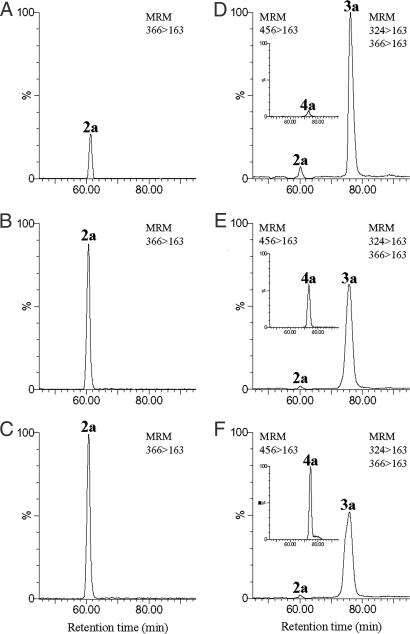
Two glycosylation steps each involving NDP sugar cosubstrates are proposed to be required for gentamicin biosynthesis: the addition of d-glucosamine (GlcN) to form the pseudodisaccharide intermediate paromamine (3a), and the subsequent addition of d-xylose to form gentamicin A2 (4a) (Fig. 1). Two glycosyltransferases genes (gtmE and gtmG) identified within the gentamicin gene cluster (12) were expressed separately in the heterologous host to determine their roles. First, pYJ498 was constructed containing gtmG together with the 2-DOS biosynthetic genes (gtmB-gtmA-gacH) plus the resistance genes (gtmF-gtmK-gtmL). However, transformants containing pYJ498 did not synthesize paromamine (3a) but instead produced ≈105 μg/liter of 2′-N-acetylparomamine (2a) (corrected for ≈40% mean recoveries of 2a) (Fig. 2D). In contrast, transformants harboring pYJ499, derived from pYJ498 by replacement of gtmG by gtmE, produced neither 2′-N-acetylparomamine (2a) nor paromamine (3a). It therefore seemed plausible that GtmG might be accepting NDP-GLcNA rather than NDP-GlcN as the cosubstrate for attachment to 2-DOS (1a). This was confirmed by using cell-free extracts from the recombinant S. venezuelae because of difficulties in obtaining soluble GtmG protein by using standard expression systems. The crude cell-free extracts of the strain harboring pYJ498 were prepared and incubated with different concentrations of exogenous UDP-GLcNA. The amount of 2′-N-acetylparomamine (2a) formed was elevated up to 4-fold as the input of UDP-GLcNA increased (Fig. 3 A–C). As expected, in vitro experiment using the same cell-free extracts supplemented with UDP-GlcN at the respective concentrations did not result in the production of paromamine (data not shown). It has been suggested by others (15) that a putative UDP-GLcNA glycosyltransferase gene, btrM (from the butirosin gene cluster) and its homologues (neo8, kanF, and gtmG from neomycin, kanamycin, and gentamicin biosynthetic gene clusters, respectively; sequence alignment of the predicted amino acid sequences of these genes is illustrated in Fig. S4) encode enzymes that catalyze the transfer of UDP-GLcNA to 2-DOS (1a) to give 2′-N-acetylparomamine (2a). Although those other enzymes have yet to be functionally characterized using purified proteins, our present results strongly suggest that GtmG is indeed a UDP-GLcNA glycosyltransferase, which is a previously uncharacterized enzyme involved in the formation of pseudodisaccharide intermediate during aminoglycoside biosynthesis.
Fig. 3.
HPLC-ESI-MS/MS chromatograms for the glycosyltransferase-catalyzed production of 2′-N-acetylparomamine (2a) and gentamicin A2 (4a). (A–C) GtmG assay using cell-free extracts from S. venezuelae strain harboring pYJ498 (A) without exogenous UDP-N-acetyl-d-glucosamine (UDP-GLcNA), (B) with addition of 0.11 mM UDP-GLcNA, and (C) with addition of 0.55 mM UDP-GLcNA. (D–F) GtmE assay using cell-free extracts from S. venezuelae strain harboring pYJ504 (D) without exogenous UDP-D-xylose (UDP-Xyl), (E) with addition of 0.12 mM UDP-Xyl, and (F) with addition of 0.60 mM UDP-Xyl. Monitoring of gentamicin A2 (4a) (456 > 163) is separately illustrated as an Inset to each chromatogram (D, E, and F).
GtmM Acts as a 2′-N-Acetylparomaine Deacetylase to Generate Paromaine (3a).
During the biosynthesis of butirosin, a paromamine deacetylase, BtrD, acts on 2′-N-acetylparomamine (2a) to produce paromamine (3a) (18), and a similar activity is required during the biosynthesis of gentamicin. A homologue of btrD is found in the region flanking either side of genD from the extended gentamicin biosynthetic gene cluster (GenBank accession no. AJ628149). Detailed sequence analysis revealed a possible start codon 142 nt upstream of genD gene. Further DNA sequencing of the PCR product of the ORF-corrected genD found the insertion of single cytosine residue at the 51th nucleotide position in genD, causing the frame shift in the coding region. A new ORF (gtmM, GenBank accession no. AM946392) encoding putative deacetylase is located 11,096 nt downstream of gacH. When the deduced product of gtmM was aligned with those of the well characterized deacetylases, two highly conserved regions (HXDD and HXDH) (18) were evident (see Fig. S5). We imported the deacetylase genes btrD, kacA, neo16, and gtmM from the gene clusters for butirosin, kanamycin, neomycin, and gentamicin biosynthesis, respectively (5, 6, 10). The four deacetylase genes were separately cloned into constructs containing the genes for 2-DOS biosynthesis, glycosyltransfer, and resistance (gtmB-gtmA-gacH-gtmF-gtmG-gtmK-gtmL) to produce plasmids pYJ500, pYJ501, pYJ502, and pYJ503, respectively. Expression of all four plasmids in the heterologous host led to production of paromamine (3a) along with its precursor 2′-N-acetylparomamine (2a) (Fig. 2E). Strains expressing the Streptomyces genes kacA, neo16, and gtmM biosynthesized similar amounts of paromamine (3a) (150 μg per liter, corrected for ≈90% mean recoveries of 3a), whereas slightly lower amounts were obtained with btrD, which is of Bacillus origin and might have been expressed less efficiently in the heterologous host, S. venezuelae. These results verify KacA, Neo16, and GtmM as deacetylases involved in the biosynthesis of paromamine (3a) and are in agreement with the previous report that paromamine (3a) is produced via the intermediate 2′-N-acetylparomamine (2a) using His6-tagged BtrD (18). Here, we produced both 2′-N-acetylparomamine (2a) and paromamine (3a) in vivo, which reveals that 2′-N-acetylparomamine (2a) is a true biosynthetic intermediate in the formation of gentamicin and other aminoglycosides.
GtmE Acts as an UDP-Xylose Glycosyltransferase to Generate Gentamicin A2 (4a).
A second glycosylation, using NDP-d-xylose (NDP-Xyl) as cosubstrate, is required for the formation of gentamicin A2 (4a) from paromamine (3a). Expression of gtmE with the genes necessary for biosynthesis of paromamine (gtmB-gtmA-gacH-gtmF-gtmG-kacA/gtmM-gtmK-gtmL) led to production of gentamicin A2 (4a) together with its precursors 2′-N-acetylparomamine (2a) and paromamine (3a) (Fig. 2F). Therefore, GtmE appears to be the glycosyltransferase required for the transfer of NDP-Xyl to paromamine (3a). Sequence alignment of the predicted product of gtmE with other putative glycosyltransferases, which are found in the biosynthetic gene clusters of other 4,6-disubstituted aminoglycosides, is shown in Fig. S6. The yield of gentamicin A2 (4a) obtained from the culture extract was only 4.5 μg/liter (corrected for ≈95% mean recoveries of 4a), and a substantial amount of paromamine (3a) remained unaltered, suggesting that NDP-Xyl might be present in S. venezuelae in limiting amounts. To determine whether d-xylose is attached directly to paromamine or whether d-xylose is generated from glucose or ribose, whereas they are bound to paromamine (26), crude cell-free extracts from the strain harboring pYJ504 were prepared and incubated with different concentrations of exogenous UDP-Xyl and UDP-glucose (UDP-Glc). Increased input of UDP-Xyl resulted in enhanced production of gentamicin A2 up to 8-fold (4a), whereas in reciprocal fashion the amount of residual paromamine (3a) dropped significantly (Fig. 3 D–F). However, UDP-Glc did not support biosynthesis of gentamicin A2. Although NDP-ribose could not be tested because of its unavailability, these results provide evidence that UDP-d-xylose (UDP-Xyl) is a cofactor for gentamicin A2 biosynthesis, and that its transfer to paromamine is catalyzed by GtmE. It was reported (27) that an UDP-glucuronic acid (UDP-GlcA) decarboxylase AviE2 was involved in the formation of UDP-Xyl in avilamycin-producing Streptomyces viridochromogenes Tu57 (27). CalS8 of calicheamicin-producing M. echinospora was reported as an UDP-Glc dehydrogenase involved in deoxypentose biosynthesis. Furthermore, several natural products such as maduropeptin from Actinomadura madurea, esperamicin from Actinomadura verrucosospora, and evernimicin from Micromonospora carbonacea bear deoxypentose(s) (28). UDP-Glc dehydrogenase and UDP-GlcA decarboxylase may be the key enzymes necessary to synthesize nucleotide-activated deoxypentoses such as UDP-Xyl in actitomycetes in a similar way to plants and fungi (29, 30), and it seems likely that such metabolic pathways are present in S. venezuelae to provide NDP-Xyl.
In conclusion we have elucidated the minimal complement of genes required for the formation of gentamicin A2 and identified its biosynthetic intermediates. 2-DOS (1a) is synthesized by the products of gtmB, gtmA, and gacH. The gtmG gene encodes a glycosyltransferase that transfers GLcNA to produce 2′-N-acetylparomamine (2a). A deacetylase GtmM required to remove the acetyl group to form paromamine (3a) was found in M. echinospora, and GtmM and its homologues BtrD, KacA, and Neo16 were each able to carry out this function. GtmE attaches d-xylose to paromamine (3a) to produce gentamicin A2 (4a). The constructed gene combinations and the obtained biosynthetic intermediates are shown in Table 1. The assigned functions of these genes are summarized in Table 2. This report represents an in vivo study regarding the expression of biosynthetic enzymes for complete synthesis of the pseudotrisaccharide form of an aminoglycoside in a heterologous host. Direct evidence is provided for the biosynthetic route to this 4,6-disubstituted aminoglycoside allowing the unequivocal assignment of specific gene products to each biosynthetic step. Further studies regarding the in vitro characterization of both glycosyltransferases, GtmG and GtmE, toward their substrates are impending. A detailed understanding of the complete biosynthetic pathway of gentamicin C components would assist in comprehending the biosynthesis of other 4,6-disubstituted aminoglycosides and form the basis for evolution of pathway engineering toward the generation of aminoglycosides active against rapidly emerging aminoglycoside-resistant pathogens. A continuing challenge of this heterologous expression approach is the relatively low productivity of the target compounds. However, understanding the limited intracellular availability of one cosubstrate (NDP-d-xylose) provided a strategy for enhancing the production level.
Table 1.
Constructed gene combinations and the products obtained
| Plasmid | Gene combination* | Product† |
|||
|---|---|---|---|---|---|
| 1a | 2a | 3a | 4a | ||
| pYJ489 | gtmF-gtmK-gtmL | Resistance genes | |||
| pYJ494 | gtmD-gtmB-gtmA-gtmC | × | × | × | × |
| pYJ495 | gtmD-gtmB-gtmA-gacH | ○ | × | × | × |
| pYJ496 | gtmB-gtmA-gacH | ○ | × | × | × |
| pYJ497 | gtmD-gtmA-gacH | × | × | × | × |
| pYJ498 | gtmB-gtmA-gacH-gtmF-gtmG-gtmK-gtmL | ○ | ○ | × | × |
| pYJ499 | gtmB-gtmA-gacH-gtmF-gtmE-gtmK-gtmL | ○ | × | × | × |
| pYJ500 | gtmB-gtmA-gacH-gtmF-gtmG-btrD-gtmK-gtmL | ○ | ○ | ○ | × |
| pYJ501 | gtmB-gtmA-gacH-gtmF-gtmG-kacA-gtmK-gtmL | ○ | ○ | ○ | × |
| pYJ502 | gtmB-gtmA-gacH-gtmF-gtmG-neo16-gtmK-gtmL | ○ | ○ | ○ | × |
| pYJ503 | gtmB-gtmA-gacH-gtmF-gtmG-gtmM-gtmK-gtmL | ○ | ○ | ○ | × |
| pYJ504 | gtmB-gtmA-gacH-gtmF-gtmG-gtmE-kacA-gtmK-gtmL | ○ | ○ | ○ | ○ |
| pYJ505 | gtmB-gtmA-gacH-gtmF-gtmG-gtmE-gtmM-gtmK-gtmL | ○ | ○ | ○ | ○ |
*Using pSE34 E. coli-Streptomyces shuttle vectors (19).
†1a, 2a, 3a, and 4a representing 2-deoxystreptamine, 2'-N-acetylparomamine, paromamine, and gentamicin A2, respectively.
Table 2.
Functional assignment of genes associate with the biosynthesis gentamicin A2
| Gene | Proposed role of the gene product | Ref(s) |
|---|---|---|
| gtmA | 2-DOI synthase | 12 |
| gtmB | 2-DOI and keto-2-DOIA aminotransferase | This work |
| gacH | 2-DOIA dehydrogenase | This work |
| gtmG | NDP-N-acetyl-D-glucosaminyl transferase | This work |
| gtmM | 2'-N-Acetylparomamine deacetylase | This work |
| gtmE | NDP-D-xylosyl transferase | This work |
| BtrD | 2'-N-Acetylparomamine deacetylase | 15 |
| kacA | 2'-N-Acetylparomamine deacetylase | This work |
| neo16 | 2'-N-Acetylparomamine deacetylase | This work |
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions.
Escherichia coli DH10B and plasmid Litmus28 (New England Biolabs) were used for routine subcloning. High-copy number E. coli-Streptomyces shuttle vector pSE34 containing the strong ermE* promoter plus a thiostrepton resistance marker was used as an expression plasmid (19). The gentamicin biosynthetic genes from M. echinospora are located on the cosmid pGEN01 (12). S. venezuelae YJ003, which is deficient in biosynthesis of the endogenous deoxysugar TDP-d-desosamine, was used as a heterologous host (20). Protoplast formation and transformation procedures of S. venezuelae were performed as described (31). S. venezuelae YJ003 and derived strains were maintained on SPA agar (20) and grown in liquid R2YE (31) at 30°C for preparation of protoplasts, which were regenerated on R2YE agar medium supplemented with thiostrepton (30 μg/ml). The E. coli strains were grown in LB medium supplemented with ampicillin (50 μg/ml) to select for plasmid vectors. For production of gentamicin intermediates, S. venezuelae strains expressing gentamicin biosynthetic genes were cultivated at 30°C for 4 days in one liter of baffled Erlenmeyer flasks containing 300 ml of R2YE medium supplemented with thiostrepton (25 μg/ml). To obtain authentic gentamicin A2, M. echinospora ATCC 15385 was grown for 7 days at 28°C in 50 ml of N-Z amine medium (1% glucose, 2% soluble starch, 0.5% yeast extract, 0.5% N-Z amine and 0.2% calcium carbonate) in 500-ml baffled flasks on a rotary shaker (25).
PCR Amplification of Gentamicin Biosynthetic Genes.
DNA fragments containing various gentamicin biosynthetic genes were amplified by PCR (Table S1). Each pair of deoxyoligonucleotide primers contained two restriction sites to facilitate subcloning of each DNA fragment. PCR was performed by using Pfu polymerase (Fermentas) under the manufacturer's recommended conditions. The DNA fragments containing each of gtmF, gtmK, gtmL, gtmD, gtmB, gtmA, gtmC, gacH, gtmG, and gtmE were amplified by using pGEN01 as a template (12). The fragments encoding GtmM, BtrD, KacA, and Neo16 were amplified by using genomic DNA from M. echinospora ATCC 15385, Bacillus circulans ATCC 21557, S. kanamyceticus ATCC 12853, and S. fradiae ATCC 10745, respectively, due to template. All PCR products were cloned into Litmus28 and sequenced to confirm their authenticity.
Construction of Expression Plasmids.
This was done as summarized in Table 1. Amplified products of the putative resistance genes gtmF-gtmK-gtmL were ligated in series into pSE34 by using standard protocols, and the resultant plasmid, pYJ489, was used to confer gentamicin resistance in S. venezuelae.
For expression of a combination of putative 2-DOS (1a) biosynthetic genes, PCR-amplified fragments containing gtmD, gtmB, gtmA, gtmC, or gacH were ligated in series into Litmus28 and then transplanted into pSE34 to generate a set of plasmids containing gtmD-gtmB-gtmA-gtmC, gtmD-gtmB-gtmA-gacH, gtmB-gtmA-gacH, and gtmD-gtmA-gacH. These were designated pYJ494, pYJ495, pYJ496, and pYJ497, respectively. In a further set of constructions, the resistance genes gtmF-gtmK-gtmL from pYJ489 were introduced into pYJ496 together with either of the glycosyltransferase genes gtmG or gtmE to generate pYJ498 or pYJ499, respectively. The latter two vectors allowed expression of 2-DOS biosynthetic genes and gentamicin resistance determinants in addition to the respective glycosyltransferase genes. The next set of expression vectors, pYJ500, pYJ501, pYJ502, and pYJ503, were equivalent to pYJ498 supplemented with deacetylase genes btrD, kacA, neo16, and gtmM, respectively, and finally introduction of gtmE into gtmG-containing pYJ501 and pYJ503 generated a plasmid, pYJ504 and pYJ505, respectively, capable of expressing the entire complement of resistance determinants plus the minimal gene set for gentamicin A2 production.
Analyses of Gentamicin Biosynthetic Intermediates.
Gentamicin biosynthetic intermediates produced by S. venezuelae strains were extracted by using the OASIS MCX (Waters) SPE cleanup procedure was done as described (25) and analyzed by HPLC-ESI-MS/MS. Analytical HPLC-ESI-MS/MS was performed on a Waters/Micromass Quattro micro/MS interface using two XTerra MS C18 (3.5 μm, Waters) columns, 150 × 2.1 and 250 × 2.1 mm, connected in tandem. The analytes were eluted at a flow rate of 60 μl/min with 30% (vol/vol) acetonitrile in water with 10 mM heptafluorobutyric acid (HFBA, Fluka) and monitored by ESI in the positive mode. Quantification of the analytes was conducted by using MS/MS in the multiple reactions monitoring mode. This was done by selecting the two mass ions set to detect a transition of the parent ion to the product ion specific to the selected analytes (26): 2-DOS (1a), 163 > 102; 2′-N-acetylparomamine (2a), 366 > 163; paromamine (3a), 324 > 163; gentamicin A2 (4a), and 456 > 163. Five separate cultivations and extractions were performed.
Isolation and Identification of Gentamicin Biosynthetic Intermediates as Authentic Standards.
2-DOS (1a) produced by S. venezuelae strains was identified by using commercially available 2-DOS (1a) (GeneChem) as a standard. 2′-N-acetylparomamine (2a) was purified directly from an S. venezuelae strain harboring the plasmid pYJ498, and its spectral data were authenticated by comparison with previous data (18). Paromamine (3a) from S. venezuelae expressing plasmid pYJ501 was identified by using standard material obtained from hydrolysis of paromomycin (Sigma). Gentamicin A2 (4a) produced by S. venezuelae harboring pYJ504 was compared with its purified form from wild-type M. echinospora. Details for the preparation and analysis of both authentic standards and gentamicin biosynthetic intermediates produced by S. venezuelae strains harboring the respective gene sets are given in SI Text; see also Figs. S7–S14 Tables S2–S5. Preparative HPLC was performed with a Spherisorb S5 ODS2 (250 × 20 mm, Waters) semiprep column eluted with the same mobile phase used in the analytical HPLC system, at a flow rate of 12 ml/min over a period of 100 min. This eluent was fractionated into 5-ml portions that were monitored by analytical HPLC-ESI-MS/MS to detect and characterize the presence of gentamicin biosynthetic intermediates. Fractions containing products of interest were pooled and extracted by using OASIS MCX SPE cleanup (25) followed by freeze-drying. 1H, 13C, and 2D 1H-1H COSY NMR spectra were acquired by using a Varian INOVA 500 spectrometer at 298 K. Chemical shifts were reported in ppm by using trimethylsilyl-2,2,3,3-tetradeuteropropionic acid (TSP) as an internal reference. The assignment of each compound was carried out by comparison with previously assigned 1H NMR spectra (18, 32–34) and by a combination of 1D and 2D NMR experiments. All NMR data processing was done by using MESTREC (Magnetic Resonance Companion) software.
In Vitro Glycosyltransfer Reactions Using Cell-Free Extracts.
Cell-free extracts of S. venezuelae strains harboring pYJ498 or pYJ504 were prepared by glass-bead homogenization (31). The mycelium (6-g wet weight) of each strain had been cultivated in R2YE medium for 3 days at 30°C, harvested by centrifugation, washed twice with 0.1 M Tris·HCl (pH 7.6), and then resuspended in 20 ml of extraction buffer (0.1 M Tris·HCl, 10 mM MgCl2, 6 mM 2-mercaptoethanol, 1 mM PMSF, pH 7.6) at 4°C. After mixing with 10 g of precooled glass beads (150 to 212 μm in diameter, Sigma), the mycelium was disrupted by vigorous agitation by using a vortex mixer. This involved 10 bursts of 30 sec each, with intermittent cooling in ice. Next, the glass beads were eliminated by low-speed centrifugation, and cellular debris was removed by centrifugation at 18,000 × g for 20 min. The entire process was carried out at 4°C and generated 15 ml of resulting supernatant. As one of proposed substrates for GtmG reaction, UDP-GlcN was prepared by enzymatic reaction of UDP-d-glucose pyrophosphorylase (E.C. 2.7.7.9, Sigma) with glucosamine-1-phosphate (Sigma) and UTP (Sigma), as described in ref. 35. Glycosyltransfer reactions were initiated by adding activated sugar cosubstrates to the cell-free extracts as follows: for GtmG, UDP-GLcNA (GeneChem) or UDP-GlcN was used at final concentrations between 0 and 0.55 mM; for GtmE, UDP-d-xylose (CarboSource Services, University of Georgia, Griffin, GA) or UDP-Glc (Sigma) was used at final concentrations between 0 and 0.60 mM. Reaction mixtures were incubated at 30°C for 10 h before quenching with 15 ml of ice-cold phenol/chloroform/isoamyl alcohol (25:24:1, Sigma). After centrifugation, gentamicin-related compounds were extracted by using OASIS MCX SPE cleanup as previously described and then reconstituted to 200 μl with water. A portion of each extract was subjected to HPLC-ESI-MS analysis (see conditions described above). Independent experiments were performed in duplicate.
Supplementary Material
Acknowledgments.
We thank Prof. Eric Cundliffe (University of Leicester, Leicester, U.K.) for helpful suggestions and critical reading of this manuscript. This work was supported by Ministry of Commerce, Industry, and Energy (MOCIE); the Marine and Extreme Genome Research Center Program of the Ministry of Land, Transportation and Maritime Affairs; Korea Science and Engineering Foundation (KOSEF), funded by the Korea government (MEST) (Grant M10749000201-07N4900-20110, R11-2005-008-00000-0) and the National Research Laboratory (NRL) program grant (R0A-2008-000-20030-0). J.W.P. gratefully acknowledges a Grant 20080401034038 from the BioGreen 21 program, Rural Development Administration, and we thank the Ministry of Education for the Brain Korea 21 Fellowship.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AM946392).
This article contains supporting information online at www.pnas.org/cgi/content/full/0803164105/DCSupplemental.
References
- 1.Weinstein MJ, Luedemann GM, Oden EM, Wagman GH. Gentamicin, a new antibiotic complex from Micromonospora. Antimicrob Agents Chemother. 1963;122:463–464. [PubMed] [Google Scholar]
- 2.Carter AP, et al. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 3.Schatz A, Bugie E, Waksman SA. Streptomycin, a substance exhibiting antibiotic activity against gram-positive and gram-negative bacteria. Proc Soc Exp Med Biol. 1944;55:66–69. doi: 10.1097/01.blo.0000175887.98112.fe. [DOI] [PubMed] [Google Scholar]
- 4.Testa RT, Tilley BC. Biotransformation, a new approach to aminoglycoside biosynthesis: II. Gentamicin. J Antibiot. 1976;29:140–146. doi: 10.7164/antibiotics.29.140. [DOI] [PubMed] [Google Scholar]
- 5.Ota Y, et al. Butirosin-biosynthetic gene cluster from Bacillus circulans. J Antibiot. 2000;53:1158–1167. doi: 10.7164/antibiotics.53.1158. [DOI] [PubMed] [Google Scholar]
- 6.Huang F, et al. The neomycin biosynthetic gene cluster of Streptomyces fradiae NCIMB 8233: Characterisation of an aminotransferase involved in the formation of 2-deoxystreptamine. Org Biomol Chem. 2005;3:1410–1418. doi: 10.1039/b501199j. [DOI] [PubMed] [Google Scholar]
- 7.Huang F, et al. Elaboration of neosamine rings in the biosynthesis of neomycin and butirosin. ChemBioChem. 2007;8:283–288. doi: 10.1002/cbic.200600371. [DOI] [PubMed] [Google Scholar]
- 8.Llewellyn NM, Li Y, Spencer JB. Biosynthesis of butirosin: transfer and deprotection of the unique amino acid side chain. Chem Biol. 2007;14:379–386. doi: 10.1016/j.chembiol.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Kudo F, Fujii T, Kinoshita S, Eguchi T. Unique O-ribosylation in the biosynthesis of butirosin. Bioorg Med Chem. 2007;15:4360–4368. doi: 10.1016/j.bmc.2007.04.040. [DOI] [PubMed] [Google Scholar]
- 10.Kharel MK, et al. A gene cluster for biosynthesis of kanamycin from Streptomyces kanamyceticus: Comparison with gentamicin biosynthetic gene cluster. Arch Biochem Biophys. 2004;429:204–214. doi: 10.1016/j.abb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Unwin J, et al. Gene cluster in Micromonospora echinospora ATCC 15835 for the biosynthesis of the gentamicin C complex. J Antibiot. 2004;57:436–445. doi: 10.7164/antibiotics.57.436. [DOI] [PubMed] [Google Scholar]
- 12.Kharel MK, et al. Molecular cloning and characterization of a 2-deoxystreptamine biosynthetic gene cluster in gentamicin-producing Micromonospora echinospora ATCC 15835. Mol Cells. 2004;18:71–78. [PubMed] [Google Scholar]
- 13.Kharel MK, et al. Isolation and characterization of the tobramycin biosynthetic gene cluster from Streptomyces tenebrarius. FEMS Microbiol Lett. 2004;230:185–190. doi: 10.1016/S0378-1097(03)00881-4. [DOI] [PubMed] [Google Scholar]
- 14.Love SF, Maiese WM, Rothstein DM. Conditions for protoplasting, regenerating, and transforming the calicheamicin producer, Micromonospora echinospora. Appl Environ Microbiol. 1992;58:1376–1378. doi: 10.1128/aem.58.4.1376-1378.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Llewellyn NM, Spencer JB. Biosynthesis of 2-deoxystreptamine-containing aminoglycoside antibiotics. Nat Prod Rep. 2006;23:864–874. doi: 10.1039/b604709m. [DOI] [PubMed] [Google Scholar]
- 16.Testa RT, Tilley BC. Biosynthesis of sisomicin and gentamicin. Jpn J Antibiot. 1979;32(Suppl):S47–S59. [PubMed] [Google Scholar]
- 17.Kase H, Odakura Y, Nakayama K. Sagamicin and the related aminoglycosides: fermentation and biosynthesis. I. Biosynthetic studies with the blocked mutants of Micromonospora sagamiensis. J Antibiot. 1982;35:1–9. doi: 10.7164/antibiotics.35.1. [DOI] [PubMed] [Google Scholar]
- 18.Truman AW, Huang F, Llewellyn NM, Spencer JB. Characterization of the enzyme BtrD from Bacillus circulans and revision of its functional assignment in the biosynthesis of butirosin. Angew Chem Int Ed. 2007;46:1462–1464. doi: 10.1002/anie.200604194. [DOI] [PubMed] [Google Scholar]
- 19.Smirnova N, Reynolds KA. Engineered fatty acid biosynthesis in Streptomyces by altered catalytic function of beta-ketoacyl-acyl carrier protein synthase III. J Bacteriol. 2001;183:2335–2342. doi: 10.1128/JB.183.7.2335-2342.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong JSJ, et al. New olivosyl derivatives of methymycin/pikromycin from an engineered strain of Streptomyces venezuelae. FEMS Microbiol Lett. 2004;238:391–399. doi: 10.1016/j.femsle.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Huang F, Li Y, Yu J, Spencer JB. Biosynthesis of aminoglycoside antibiotics: Cloning, expression and characterisation of an aminotransferase involved in the pathway to 2-deoxystreptamine. Chem Commun. 2002;7:2860–2861. doi: 10.1039/b209799k. [DOI] [PubMed] [Google Scholar]
- 22.Kudo F, et al. Biosynthesis of 2-deoxystreptamine by three crucial enzymes in Streptomyces fradiae NBRC 12773. J Antibiot. 2005;58:766–774. doi: 10.1038/ja.2005.104. [DOI] [PubMed] [Google Scholar]
- 23.Yokoyama K, et al. Stereochemical recognition of doubly functional aminotransferase in 2-deoxystreptamine biosynthesis. J Am Chem Soc. 2005;127:5869–5874. doi: 10.1021/ja0445948. [DOI] [PubMed] [Google Scholar]
- 24.Yokoyama K, et al. Characterization and mechanistic study of a radical SAM dehydrogenase in the biosynthesis of butirosin. J Am Chem Soc. 2007;129:15147–15155. doi: 10.1021/ja072481t. [DOI] [PubMed] [Google Scholar]
- 25.Park JW, et al. Analytical profiling of biosynthetic intermediates involved in the gentamicin pathway of Micromonospora echinospora by high-performance liquid chromatography using electrospray ionization mass spectrometric detection. Anal Chem. 2007;79:4860–4869. doi: 10.1021/ac070028u. [DOI] [PubMed] [Google Scholar]
- 26.Rinehart KL., Jr Biosynthesis and mutasynthesis of aminocyclitol antibiotics. Jpn J Antibiot. 1979;32(Suppl):S32–S46. [PubMed] [Google Scholar]
- 27.Hofmann C, et al. Genes encoding enzymes responsible for biosynthesis of L-lyxose and attachment of eurekanate during avilamycin biosynthesis. Chem Biol. 2005;12:1137–1143. doi: 10.1016/j.chembiol.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 28.Bililign T, Shepard EM, Ahlert J, Thorson JS. On the origin of deoxypentoses: Evidence to support a glucose progenitor in the biosynthesis of calicheamicin. ChemBioChem. 2002;3:1143–1146. doi: 10.1002/1439-7633(20021104)3:11<1143::AID-CBIC1143>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 29.Harper AD, Bar-Peled M. Biosynthesis of UDP-xylose. Cloning and characterization of a novel Arabidopsis gene family, UXS, encoding soluble and putative membrane-bound UDP-glucuronic acid decarboxylase isoforms. Plant Physiol. 2002;130:2188–2198. doi: 10.1104/pp.009654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bar-Peled M, Griffith CL, Doering TL. Functional cloning and characterization of a UDP-glucuronic acid decarboxylase: The pathogenic fungus Cryptococcus neoformans elucidates UDP-xylose synthesis. Proc Natl Acad Sci USA. 2001;98:12003–12008. doi: 10.1073/pnas.211229198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kieser T, et al. Practical Streptomyces Genetics. Norwich, UK: John Innes Foundation; 2000. pp. 75–97. [Google Scholar]
- 32.Nagabhushan TL, Turner WN, Daniels PJL, Morton JB. The gentamicin antibiotics. 7. Structures of the gentamicin antibiotics A1, A3, and A4. J Org Chem. 1975;40:2830–2834. doi: 10.1021/jo00907a028. [DOI] [PubMed] [Google Scholar]
- 33.Nagabhushan TL, Daniels PJL, Jaret RS, Morton JB. The gentamicin antibiotics. 8. Structure of gentamicin A2. J Org Chem. 1975;40:2835–2836. doi: 10.1021/jo00907a029. [DOI] [PubMed] [Google Scholar]
- 34.Haskell TH, French JC, Bartz QR. Paromomycin. I. Paromamine, a glycoside of D-glucosamine. J Am Chem Soc. 1959;81:3480–3481. [Google Scholar]
- 35.Lee HC, Lee SD, Sohng JK, Liou K. One-pot enzymatic synthesis of UDP-D-glucose from UMP and glucose-1-phosphate using an ATP regeneration system. J Biochem Mol Biol. 2004;37:503–506. doi: 10.5483/bmbrep.2004.37.4.503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.