Abstract
Biochemically active wheat thioredoxin h has been overexpressed in the endosperm of transgenic barley grain. Two DNA constructs containing the wheat thioredoxin h gene (wtrxh) were used for transformation; each contained wtrxh fused to an endosperm-specific B1-hordein promoter either with or without a signal peptide sequence for targeting to the protein body. Twenty-two stable, independently transformed regenerable lines were obtained by selecting with the herbicide bialaphos to test for the presence of the bar herbicide resistance gene on a cotransformed plasmid; all were positive for this gene. The presence of wtrxh was confirmed in 20 lines by PCR analysis, and the identity and level of expression of wheat thioredoxin h was assessed by immunoblots. Although levels varied among the different transgenic events, wheat thioredoxin h was consistently highly expressed (up to 30-fold) in the transgenic grain. Transgenic lines transformed with the B1-hordein promoter with a signal peptide sequence produced a higher level of wheat thioredoxin h on average than those without a signal sequence. The overexpression of thioredoxin h in the endosperm of germinated grain effected up to a 4-fold increase in the activity of the starch debranching enzyme, pullulanase (limit dextrinase), the enzyme that specifically cleaves α-1,6 linkages in starch. These results raise the question of how thioredoxin h enhances the activity of pullulanase because it was found that the inhibitor had become inactive before the enzyme showed appreciable activity.
There is a growing body of evidence that, as in photosynthesis, thioredoxin plays a primary role in regulating heterotrophic processes in plants. In this capacity, the disulfide group of a thioredoxin of the h-type is reduced by NADPH via the flavin enzyme, NADP-thioredoxin reductase (NTR) (1, 2) (Eq. 1).
 |
1 |
Biochemical studies initiated 10 years ago with wheat have provided evidence that thioredoxin h functions in germination and seedling development (3). Analysis of these results suggests that thioredoxin h, reduced via NTR with metabolically generated NADPH, acts early in seedling growth to initiate the mobilization of nitrogen and carbon in the endosperm, the major repository of storage protein and carbohydrate in cereals (4, 5). The NADPH needed for this reduction can be generated enzymatically from carbohydrate stored in the endosperm via glucose 6-phosphate and 6-phosphogluconate dehydrogenases (5).
Through the reduction of intramolecular disulfide bonds of target proteins (Eq. 2), thioredoxin h was shown to promote the degradation of major storage proteins, the inactivation of small proteins that inhibit amylolytic enzymes, and the activation of a novel calcium-dependent substrate-specific protease (4, 6, 7).
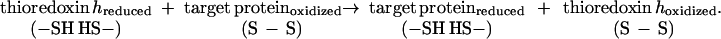 |
2 |
To gain further information on its role, we initiated a study to determine the effects of overexpressed thioredoxin h on transformed cereal grains. We now report the transformation of barley by using a wheat thioredoxin h gene (wtrxh) driven by a seed-specific promoter to target expression of the gene product to the endosperm. Stable homozygous lines overexpressing biochemically active wheat thioredoxin h have been shown to have increased activity of a starch debranching enzyme of the endosperm, pullulanase (limit dextrinase), which specifically hydrolyzes α-1,6-linkages in starch (amylopectin) during germination and seedling development (Eq. 3).
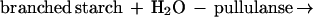 |
3 |
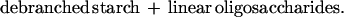 |
Thioredoxin was earlier found to increase the activity of pullulanase in vitro by inactivating a small disulfide inhibitor protein through reduction of its disulfide bonds (8). The present results suggest that thioredoxin can increase pullulanase activity in vivo in another manner, yet to be defined.
Materials and Methods
Materials.
Barley (Hordeum vulgare L.) cv. Golden Promise was used throughout this study. Chemicals and biochemicals were obtained from commercial sources and were of the highest quality available.
Construction of Wheat Thioredoxin h Expression Vectors.
For transformation, two expression vectors were constructed that contain wtrxh from bread wheat (Triticum aestivum, cv. Capitole) driven by the barley endosperm-specific B1-hordein promoter. The plasmids were constructed as follows (see Fig. 1).
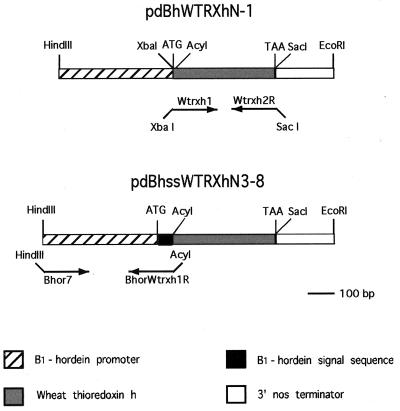
Figure 1.
Thioredoxin h constructs used for barley transformation. (Upper) pdBhWTRXhN-1 contains wtrxh under control of the barley endosperm-specific B1-hordein promoter without its signal peptide sequence and terminated by nos. (Lower) pdBhssWTRXhN3–8 contains wtrxh under control of the B1-hordein promoter with its signal peptide sequence and terminated by nos. Nucleotide sequences of the PCR-amplified wheat thioredoxin h coding region and B1-hordein 5′ flanking region were determined by DNA sequencing.
Plasmid 1 (pdBhWTRXhN-1).
A 384-bp wtrxh coding region was amplified from pTaM13.38 (9), which contains a cDNA of wtrxh that served as a template in a PCR to create XbaI and SacI sites by using the following primers: Wtrxh1 (5′-atatctagaATGGCGGCGTCGGCGGCGA-3′) and Wtrxh2R (5′-atagagctcTTACTGGGCCGCGTGTAG-3′), (see below) (small, italicized letters in the primer denote a restriction enzyme site for subcloning of the DNA fragment containing wtrxh; underlined letters denote wtrxh sequences). The ATG initiation codon for wtrxh expression was included in the Wtrxh1 primer. PCRs were performed on a thermocycler (MJ Research, Watertown, MA) by using 5 units of recombinant Taq DNA polymerase (Promega) in a 100-μl reaction volume. The reaction buffer contained 10 mM Tris⋅HCl (pH 9.0), 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, and 50 μM of each deoxyribonucleoside triphosphate. PCR conditions used 25 cycles at 94°C for 1 min, at 55°C for 1 min, and at 72°C for 2 min, with a final extension step at 72°C for 7 min. The wtrxh fragment was purified from a 0.7% agarose gel by using a QIAquick gel extraction kit (Qiagen, Chatsworth, CA), digested with XbaI and SacI and ligated into XbaI/SacI-digested pUC19 to generate the pWTRXh-1 plasmid. Nucleotide sequences of the PCR-amplified wtrxh coding region fragment were determined by the dideoxynucleotide chain termination method (United States Biochemical) with double-stranded plasmid templates and regularly spaced primers. pdBhWTRXhN-1 was made by replacing the gene encoding β-glucuronidase (uidA) in pdBhGN-1 (B1-hordein promoter-uidA-nos 3′ terminator, where nos is the Agrobacterium tumefaciens nopaline synthase 3′ polyadenylation signal; M.-J.C., unpublished work) with the XbaI/SacI fragment containing the wtrxh coding sequence from pWTRXh-1.
Plasmid 2 (pdBhssWTRXhN3–8).
Primers Bhor7 (5′-GTAAAGCTTTAACAACCCACACATTG-3′) and BhorWtrxh1R (5′-CCGACGCCGCTGCAATCGTACTTGTTGCCGCAAT-3′) containing HindIII and AcyI sites, respectively (italic letters denote restriction enzyme sites) were used for amplification of a 0.49-kb B1-hordein 5′ region, which included the B1-hordein signal peptide sequence from plasmid λ2–4/HindIII, which contains a genomic clone of B1-hordein (10). The primer BhorWtrxh1R is an overlapping primer, which contains a part of the wtrxh coding sequence (underlined) and a partial signal peptide sequence (doubly underlined) from the B1-hordein promoter, but lacks the ATG initiation codon for wtrxh. pdBhssWTRXhN3–8 was made by replacing the B1-hordein promoter in pdBhWTRXhN-1 with the 0.49-kb PCR-amplified HindIII/AcyI fragment. This construct pdBhssWTRXhN3–8 contains the barley endosperm-specific B1-hordein promoter with its signal peptide sequence, wtrxh and nos (see Fig. 1). The signal peptide sequence containing the ATG initiation codon was ligated directly to the sequence of wtrxh, with no extra amino acid sequences being introduced between the two. The authenticity of the PCR-amplified fragment from the chimeric product was confirmed by DNA sequencing.
Stable Transformation.
Stable transgenic lines of barley expressing wheat thioredoxin h driven by the B1-hordein promoter with or without the signal peptide sequence were obtained after modifications of published protocols (11, 12). Whole immature barley embryos (1.0–2.5 mm) were aseptically removed, placed scutellum-side down on DC callus-induction medium containing 2.5 mg/liter 2,4-D and 5.0 μM CuSO4 (13). One day after incubation at 24 ± 1°C in the dark, the immature embryos were transferred scutellum-side up to 2,4-dichlorophenoxyacetic acid-copper (DC) medium containing equimolar amounts of mannitol and sorbitol to give a final concentration of 0.4 M. Four hours after initiation of treatment with the osmoticum, the immature embryos were used for bombardment. Gold particles (1.0 μm) were coated with 25 μg of a 1:1 molar ratio of pAHC20 containing the bar gene under the control of the maize ubiquitin promoter (14) and one of two plasmids, pdBhWTRXhN-1 or pdBhssWTRXhN3–8. The microprojectiles were bombarded by using a PDS-1000 He biolistic device (Bio-Rad) at 1,100 psi; osmoticum treatment continued until 16–18 hr postbombardment. Bombarded immature embryos were selected on DC medium with 5 mg/liter bialaphos (Meiji Seika, Yokohama, Japan) for 2–3 months, subculturing every 2–3 weeks. Bialaphos-resistant callus was transferred onto an intermediate culturing medium (DBC2) (13), containing 2.5 mg/liter 2,4-D, 0.1 mg/liter 6-benzylaminopurine, 5.0 μM CuSO4, and 5 mg/liter bialaphos before the regeneration step (FHG medium) (15). The intermediate culturing step was carried out under dim light conditions (approximately 10–30 μE, 16 hr light) for 1–2 months. Regenerated shoots were transferred to Magenta boxes containing rooting medium (callus-induction medium without phytohormones) containing 3 mg/liter bialaphos. When shoots reached the top of the box, plantlets were transferred to soil in the greenhouse.
Cytological Analysis.
Cytological analysis of transgenic barley plants was performed as described (16) by using healthy root tips collected from young plants grown in the greenhouse.
Herbicide Application.
To determine herbicide sensitivity of T0 plants and their progeny, a section of a leaf blade at the 4- to 5-leaf stage was painted by using a cotton swab soaked in 0.25% (vol/vol) Basta solution (starting concentration 200 g/liter phosphinothricin, Hoechst Pharmaceuticals) plus 0.1% Tween 20 (17). Plants were scored 1 week after herbicide application.
PCR Screening of Transgenic Lines.
Total genomic DNA from leaf tissue was purified as described (18). To test for the presence of wtrxh in genomic DNA of putatively transformed lines, 250 ng of genomic DNA was amplified by PCR using one of two primer sets unique to wtrxh, Wtrxh1 plus Wtrxh2R or Wtrxh4 (5′-CCAAGAAGTTCCCAGCTGC-3′) plus Wtrxh5R (5′-ATAGCTGCGACAACCCTGTCCTT-3′). The presence of bar was determined by using the primer set, BAR5F (5′-CATCGAGACAAGCACGGTCAACTTC-3′) and BAR1R (5′-ATATCCGAGCGCCTCGTGCATGCG-3′) (12). Amplifications were performed with Taq DNA polymerase in a 25-μl reaction (13). Twenty-five microliters of the PCR product with loading dye was subjected to electrophoresis in a 1.0% agarose gel with ethidium bromide and photographed by using exposure to UV light. Presence of 0.4-kb (Wtrxh1, Wtrxh2R) and 0.14-kb (Wtrxh4, Wtrxh5R) fragments was consistent with intact and partial wtrxh fragments, respectively; an internal 0.34-kb fragment was produced with bar primers. Putative homozygous wtrxh and null segregant lines for wtrxh were identified by segregation analysis using PCR in the T1, T2, or T3 generations.
Western Blot Analysis.
Western blot analysis was performed on clarified protein extracts prepared from selected transgenic and null segregant lines and subjected to SDS/PAGE on 12–17% polyacrylamide gradient gels at pH 8.5 at 15 mA (19). An equal amount of protein (40 μg) was diluted 1:2 (vol/vol) in Laemmli sample buffer, boiled for 3 min, and loaded onto the above gels. Proteins were transferred to nitrocellulose at a constant voltage of 40 V for 4 hr at 4°C by using a Hoefer Transphor Transfer Unit. Nitrocellulose was blocked with 5% powdered milk in buffer (20 mM Tris⋅HCl, pH 7.5, supplemented with 0.15 M NaCl) for 2 hr at 25°C, incubated in primary antibody for 4 hr at 25°C and in secondary antibody for 1 hr at 25°C. Primary antibody was wheat anti-thioredoxin h II (20) diluted 1:500; secondary antibody was goat anti-rabbit alkaline phosphatase (Bio-Rad) diluted 1:3,000. Blots were developed in nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate alkaline phosphatase color reagent (according to Bio-Rad instructions); gels were stained with Coomassie blue to confirm transfer.
Protein Determination and Image Scanning.
Protein was estimated according to Bradford (21) by using Coomassie blue reagents from Bio-Rad and γ-globulin as standard. The blots and pullulanase activity gels were scanned by using a Bio-Rad Gel Doc 1000 and analyzed by using Bio-Rad multi analyst, version 1.0.2.
Preparation of Extracts from Dry Grain.
Grain extract.
Fifteen grams of mature grain from selected heterozygous and homozygous transgenic lines (see Table 1) was ground to powder in an electric coffee grinder (about 30 sec) and extracted with 80 ml (1:5 wt/vol) of buffer (50 mM Tris⋅HCl buffer, pH 7.9, containing 1 mM EDTA, 0.5 mM PMSF, 2 mM e-amino-n caproic acid, 2 mM benzamidine-HCl) by stirring for 3 hr at 4°C. The slurry suspension was centrifuged at 25,400 × g for 20 min, and the supernatant solution was saved; pellets were re-extracted once. The combined supernatant fractions were subjected to acidification, neutralization, and (NH4)2SO4 fractionation (22). The resulting supernatant solution from the 30–90% (NH4)2SO4 fraction was concentrated by dialysis against sucrose at 4°C and subjected either to column chromatography or heat treatment.
Table 1.
Barley lines transformed with wtrxh
| Plasmids for bombardment | Transgenic barley line | DNA PCR (T0 leaf)
|
TRXh expression in T1 seeds | Ploidy | |
|---|---|---|---|---|---|
| bar | wtrx | ||||
| pdBhWTRXhN-1 + pAHC20 | GPdBhBarWtrx-1 | + | + | n.d. | Tetraploid |
| GPdBhBarWtrx-2 | + | + | + | Tetraploid | |
| GPdBhBarWtrx-3 | + | + | + | Diploid | |
| GPdBhBarWtrx-5* | + | + | n.d. | Tetraploid | |
| GPdBhBarWtrx-16 | + | − | n.d. | Tetraploid | |
| GPdBhBarWtrx-17 | + | + | n.d. | Tetraploid | |
| GPdBhBarWtrx-19 | + | + | + | Diploid | |
| GPdBhBarWtrx-20 | + | + | + | Diploid | |
| GPdBhBarWtrx-22 | + | + | + | Diploid | |
| GPdBhBarWtrx-23 | + | + | + | Diploid | |
| pdBhssWTRXhN3-8 + pAHC20 | GPdBhssBarWtrx-1 | + | − | − | Diploid |
| GPdBhssBarWtrx-2† | + | + | + | Diploid | |
| GPdBhssBarWtrx-3 | + | + | − | Diploid | |
| GPdBhssBarWtrx-7 | + | + | + | Diploid | |
| GPdBhssBarWtrx-9 | + | + | n.d. | Tetraploid | |
| GPdBhssBarWtrx-11 | + | + | − | Diploid | |
| GPdBhssBarWtrx-13 | + | + | + | Tetraploid | |
| GPdBhssBarWtrx-14 | + | + | + | Diploid | |
| GPdBhssBarWtrx-20 | + | + | + | Tetraploid | |
| GPdBhssBarWtrx-21* | + | + | n.d. | Tetraploid | |
| GPdBhssBarWtrx-22 | + | + | + | Tetraploid | |
| GPdBhssBarWtrx-29† | + | + | + | Diploid | |
n.d., not determined.
Sterile.
† Homozygous plants were obtained from these lines.
Column chromatography.
The clarified and concentrated sample was applied to a Sephadex G-50 superfine column (2.5 × 90 cm; ≈400 ml bed volume) pre-equilibrated and eluted with 30 mM Tris⋅HCl, pH 7.9, 200 mM NaCl) by using a peristaltic pump at a flow rate of 0.5 ml/min. One hundred fifty drop-fractions were collected. Selected fractions were used to measure A280nm with a Pharmacia Biotech Ultrospec 4000 and to assay for thioredoxin h activity (see below). Active fractions were pooled and stored at 4°C.
Heat treatment.
Extract was clarified by centrifugation as above. A 20-ml aliquot of the clarified extract was placed in a 65°C water-bath until the sample temperature reached (≈10 min) and was held at 60 ± 1°C for 10 additional min; samples were cooled and then centrifuged (as above). Pellets were discarded, and the supernatant solutions were concentrated by sucrose as above and stored at −20°C until use.
Germination of Barley Grain.
Grain was germinated in a dark chamber and retained for up to 5 days at 25°C under the conditions described earlier (4, 5). A set of plates from each line was removed for extract preparation each day.
Preparation of Endosperm Extracts from Imbibed Grain.
Cell-free extracts were prepared from lots of 10–20 germinated grains of equivalent root and coleoptile length within a given cohort. Endosperm was separated from the embryo and other tissues and added to Tris⋅HCl buffer (50 mM, pH 7.9) supplemented with 1 mM EDTA and 0.5 mM PMSF (1:3 to 1:6, wt/vol ratio of tissue to buffer depending on developmental stage). After grinding in a mortar on ice, the sample was clarified by centrifugation (10 min at 24,000 × g); the supernatant fraction was recovered and stored in 0.5-ml aliquots at −80°C for pullulanase assay or gel analysis.
Assay for Thioredoxin h.
Thioredoxin h was assayed as described (23). Fifty to 120 microliters of extract was preincubated with DTT, and 0.16–0.32 μl of the preincubation mixture was used for the NADP-malate dehydrogenase (MDH) assay. Control assays were conducted on identical fractions in the absence of NADP-MDH.
Assay of Pullulanase and Pullulanase Inhibitor.
Pullulanase activity was determined spectrophotometrically at 37°C by measuring dye released after 30 min at 534 nm by using red pullulan (Megazyme, Bray, Ireland) as substrate in 50 mM citrate-phosphate buffer (pH 5.2) (24). Pullulanase also was assayed on native activity gels of 7.5% acrylamide, 1.5 mm thickness, containing 1% red pullulan (25). Pullulanase inhibitor activity was determined on fractions heated to inactivate pullulanase (70°C for 15 min) by measuring the inhibition of the fractions on added purified barley malt pullulanase. The endogenous pullulanase activity was shown to be completely eliminated by this heat treatment whereas inhibitor activity was not affected (26, 27).
Results and Discussion
Transformation and Analysis of T0 Plants and Their Progeny.
The two gene constructs, when stably introduced into barley, led to expression of wheat thioredoxin h in the grain. In both cases wtrxh was driven by the B1-hordein promoter: pdBhssWTRXhN3–8 with a signal peptide sequence and pdBhWTRXhN-1 without (Fig. 1). Twenty-two independent putatively transformed, regenerable lines were identified after bialaphos selection (Table 1). Of these, all yielded bar fragments from T0 leaf tissue by PCR and all but two produced fragments specific for wtrxh, giving a 91% cotransformation frequency. Nine lines were transformed with pdBhWTRXhN-1 containing no signal sequence and 11 with the signal sequence-containing plasmid, pdBhssWTRXhN3–8. Of the 22 independent transgenic lines examined, 12 showed a normal diploid chromosome complement (2n = 2x = 14), whereas the remaining 10 were tetraploid (2n = 4x = 28) (Table 1). Grain of T1 plants and their progeny from selected wtrxh-positive lines were planted to screen for homozygous lines. Homozygous lines and null segregants were obtained from lines GPdBhssBarWtrx-2 and GPdBhssBarWtrx-29.
Characterization of Wheat Thioredoxin h Produced in Transgenic Grain.
Of the stably transformed lines that expressed wheat thioredoxin h, on average, its level was found to be higher in transformants that had the signal peptide-containing construct (Fig. 2, Table 2) than to those that did not; further analyses therefore were carried out with the lines that had the signal peptide (see also Table 3). Western blot analysis of soluble protein fractions from heterozygous mixtures of seeds from three of these lines, GPdBhssBarWtrx-7, GPdBhssBarWtrx-22, and GPdBhssBarWtrx-29 showed 5.5 times, 22 times, and 10 times more thioredoxin h, respectively, than nontransformed control grain (Table 2). The thioredoxin content of the null segregant (GPdBhssBarWtrx-29–11–10) was similar to that of the corresponding, nontransformed control.
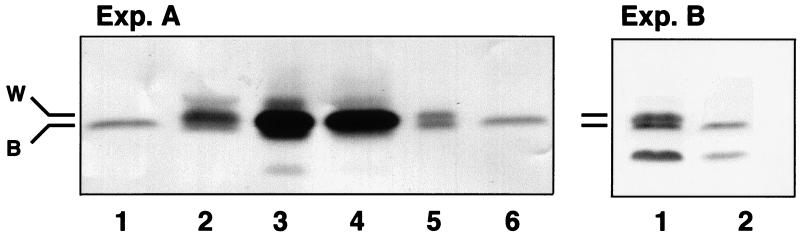
Figure 2.
Western blot analysis of extracts from segregating T1 barley grain of stable transformants containing wtrxh. Forty micrograms of soluble protein extracted from 10–20 dry grains of each line was subjected to SDS/PAGE. Western blotting was performed with wheat thioredoxin h antibody. Exp. A: lanes 1 and 6, control barley extract (cv. Golden Promise); lane 2, bread wheat extract (Triticum aestivum, cv. Capitole); lane 3, extract from GPdBhssBarWtrx-22; lane 4, extract from GPdBhssBarWtrx-29; lane 5, extract from GPdBhBarWtrx-2. Exp. B: lane 1, GPdBhBarWtrx-2; lane 2, control barley extract. W, wheat; B, barley.
Table 2.
Western blot analysis of overexpression of wheat thioredoxin h in transgenic barley grain
| Transgenic barley line | Fold overexpression |
|---|---|
| Nontransformed control | |
| GP 4-96 | 1.0* |
| Transformant with wtrxh† | |
| GPdBhBarWtrx-2 | 6.4 |
| Transformants with wtrxh and signal sequence‡ | |
| GPdBhssBarWtrx-7 | 5.5 |
| GPdBhssBarWtrx-22 | 22.0 |
| GPdBhssBarWtrx-29 | 10.0 |
| Null segregant§ | |
| GPdBhssBarWtrx-29-11-10 | 0.64 |
Corresponds to 1.46% of total volume scanned.
† pdBhWTRXhN-1.
‡ pdBhssWTRXhN3-8.
§ Originated from GPdBhssBarWtrx-29.
Table 3.
Relative total thioredoxin h activity in different transgenic barley lines
| Transgenic barley line | % total activity |
|---|---|
| Nontransformed control | |
| GP 4-96§ | 100* |
| Transformant with bar only | |
| GPBar-1 | 120 |
| Transformants with wtrxh† | |
| GPdBhBarWtrx-1 | 192 |
| GPdBhBarWtrx-22 | 151 |
| GPdBhBarWtrx-23 | 180 |
| Transformants with wtrxh and signal sequence‡ | |
| GPdBhssBarWtrx-2 | 1,650 |
| GPdBhssBarWtrx-14 | 1,723 |
| GPdBhssBarWtrx-20 | 440 |
| GPdBhssBarWtrx-22§ | 3,470 |
| GPdBhssBarWtrx-29§ | 1,316 |
Corresponds to 157 nmol NADPH oxidized per min.
† pdBhWTRXhN-1.
‡ pdBhssWTRXhN3-8.
§ Also analyzed by Western blot (compare with Table 2).
When probed on Western blots, extracts from barley typically showed one immunologically reactive band (identified by B in Fig. 2, Exp. A, lanes 1 and 6), but in some transfers showed a second faint, faster moving band (Fig. 2, Exp. B, lane 2). Tissues from transgenic lines overexpressing wtrxh were characterized by a band that did not correspond to either of the two counterparts in barley, but rather to thioredoxin h from wheat. The difference between the overexpressed 13.5-kDa wheat and the endogenous 13.1-kDa barley thioredoxin h is particularly obvious in the barley line transformed with the nontargeted thioredoxin h gene (Fig. 2, Exp. A, lane 5 and Exp. B, lane 1). Repeated analyses of the various transgenic lines by SDS/PAGE led to the conclusion that the band identified in Fig. 2 by W corresponds to the bread wheat thioredoxin introduced in barley. Independent biochemical assays with 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) (22) confirmed the ability of barley NTR to reduce wheat thioredoxin h (B. C. Yee and B.B.B., unpublished results).
Activity of Wheat Thioredoxin h Overexpressed in Barley Grain.
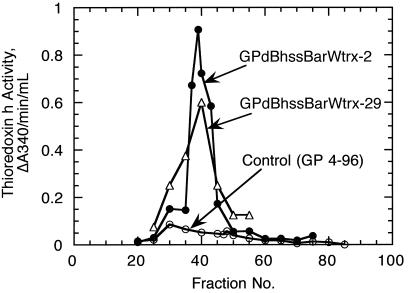
Activity of thioredoxin h in grain extracts was measured by using an assay in which thioredoxin, reduced by DTT, activates chloroplast NADP-MDH. Because of interference from other endogenous enzymes that oxidize NADPH, thioredoxin h cannot be accurately assayed in crude grain extracts, thereby necessitating a partial purification. In our initial analyses, extracts prepared from transgenic and control grain were partially purified by (NH4)2SO4 fractionation and gel filtration chromatography. Activity profiles based on these assays showed that the activity of thioredoxin h in the grain transformed with the signal sequence was much higher than that of the nontransformed control (Fig. 3).
Figure 3.
Sephadex G-50 profile of thioredoxin h activity in transgenic barley grains. Thioredoxin h activity was determined by using the NADP-MDH assay. GPdBhssBarWtrx-2 and GPdBhssBarWtrx-29 are lines stably transformed with wtrxh driven by the B1-hordein promoter with its signal peptide sequence.
Because of the tediousness of the purification protocol and the requirement for large grain quantities, the isolation procedure was modified to replace the (NH4)2SO4 and Sephadex G-50 column steps with a heat treatment. A heat step, the suitability of which was based on the early observation that Escherichia coli thioredoxin is stable at 60°C for 10 min (28), was found to decrease endogenous, nonspecific NADPH oxidation, thereby increasing the reliability of the activity measurements. Grain from nine different barley lines (transformed and nontransformed) was extracted, heat-treated, and assayed by using the NADP-MDH assay (Table 3). Total thioredoxin h activity in grain from lines segregating for the B1-hordein promoter-signal sequence construct ranged from 4- to 30-fold higher than from nontransformed lines or those transformed only with bar. Although wheat thioredoxin h appears to be relatively heat-resistant, it is not known whether all activity in the different transformed barley grain survives this treatment.
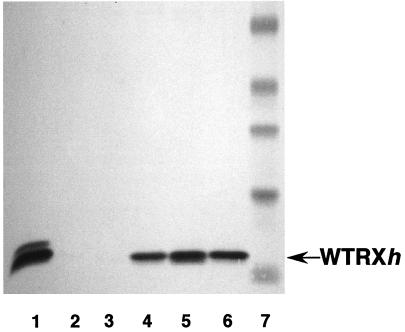
Identification of Homozygous Lines.
Because of their value in assessing biochemical attributes of the grain, homozygous wtrxh lines were identified and analyzed by Western blot. The two lines identified as homozygous both showed enhanced expression of thioredoxin h relative to that of their heterozygous parents and nontransformed controls; analysis of GPdBhssBarWtrx-29–3 is shown in Fig. 4. It is noted that demonstration of the thioredoxin h present in the nontransgenic control and null segregant grains (not obvious in the exposure shown in Fig. 4) required conditions that led to overexposure of the enriched transgenic preparations. Thioredoxin in the parent heterozygous grain was shown to be biochemically active (Fig. 3).
Figure 4.
Western blot analysis of extracts of T2 and T3 barley grain transformed with wtrxh. Forty micrograms of soluble proteins extracted from 10–20 grains of each line was fractionated by SDS/PAGE. Lane 1, wheat germ thioredoxin h; lane 2, nontransgenic control of GP 4–96; lane 3, null segregant T3 grain of GPdBhssBarWtrx-29–11–10; lane 4, heterozygous T1 grain of GPdBhssBarWtrx-29; lane 5, homozygous T2 grain of GPdBhssBarWtrx-29–3; lane 6, homozygous T3 grain of GPdBhssBarWtrx-29–3-2; lane 7, prestained standards: aprotinin, 6.9 kDa; lysozyme, 17.8 kDA; soybean trypsin inhibitor, 30.6 kDA; carbonic anhydrase, 41.8 kDa; BSA, 7l kDa.
Activity of Pullulanase.
Pullulanase is an amylolytic enzyme present in cereal grain, which has a disulfide inhibitor protein (26, 27), the activity of which is linked to thioredoxin (8). Thioredoxin, reduced by NADPH via NTR, reduces the disulfide bonds of the inhibitor, allowing the targeted pullulanase enzyme to be active. Because of this link, it was of interest to determine the activity of pullulanase in the thioredoxin h-overexpressing transformants.
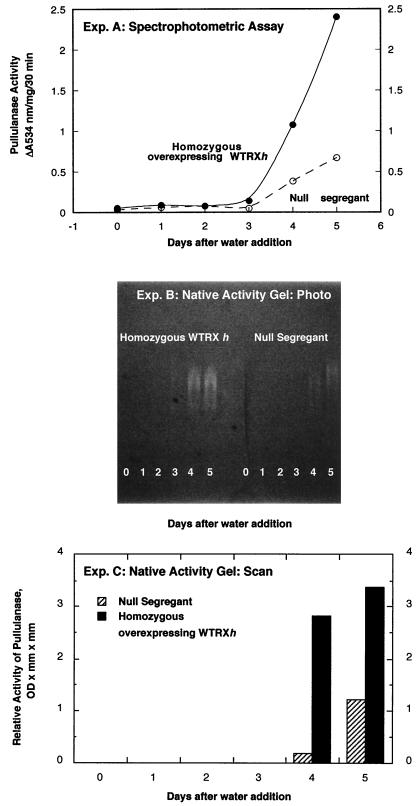
As demonstrated in spectrophotometric assays (Fig. 5 Top), extracts from transformed grain of a homozygous line (GPdBhssBarWtrx-29–3) overexpressing thioredoxin h showed a 3- to 4-fold increase in pullulanase activity on the fifth day after initiation of germination relative to a null segregant. Confirmatory results were obtained in a separate experiment with native activity gels. The increase in activity was apparent either when gels were viewed directly (Fig. 5 Middle) or when the activity on the gels was assessed by scanning and integrating the clarified bands (Fig. 5 Bottom). A homozygous line isolated from a different, independent transformation event (GPdBssBarWtrx-2–1-15) showed a similar response (data not shown). Analysis of comparable grain extracts revealed that the pullulanase inhibitor was inactive on the fourth and fifth days after water addition in both the transformant and null segregants. These results thus demonstrate that the increase in pullulanase activity observed after the third day is not caused by enhanced inactivation of the inhibitor in the transgenic grain but to a mechanism yet to be defined. It is possible that thioredoxin acts either by increasing the de novo synthesis of pullulanase (29) or by lowering the binding of the mature enzyme to the starchy endosperm. There is evidence that some of the pullulanase of the mature endosperm is present in bound form and can be solubilized by reducing conditions (30, 31).
Figure 5.
Effect of overexpressed thioredoxin h on pullulanase activity in transgenic barley grain during germination and seedling development. A homozygous line, GPdBhssBarWtrx-29–3, and a null segregant, GPdBhssBarWtrx-29–11–10, were used for the pullulanase assays. (Top) Pullulanase was assayed spectrophotometrically by measuring the dye released from red pullulan substrate at 534 nm. (Middle) Pullulanase was separated on native 7.5% polyacrylamide gels containing the red pullulan substrate. Activity, identified by comparison with purified barley pullulanase, is seen as clear areas that developed on incubating the gel in 0.2 M succinate buffer, pH 6.0, for 1 hr at 37°C. (Bottom) The gel in Exp. B was scanned and analyzed by integration of the activity bands.
Concluding Comments.
The present findings add a dimension to the role of thioredoxin h in germination and seedling development. When overexpressed and targeted to the barley grain endosperm, thioredoxin h acts to enhance pullulanase activity mainly via a mechanism independent of its earlier identified ability to inactivate a specific disulfide inhibitor protein. Next it should be elucidated how thioredoxin h mediates this effect and whether other enzymes functional during and after germination show a similar response. It will be of interest to know how this activity relates to other roles thioredoxin h is known to play in plants—for example, involvement in cell division, DNA replication, oxidative stress, and self-incompatibility (3, 32).
Acknowledgments
We thank Dr. M. B. Sørensen (Carlsberg Research Laboratory, Copenhagen, Denmark) for the gift of λ2–4/HindIII, Dr. P. Joudrier (Institut National de la Recherche Agronomique, Montpellier, France) for pTaM13.38, and Dr. P. Quail (U.S. Department of Agriculture, Plant Gene Expression Center, Albany, CA) for pAHC20. The assistance of B. C. Yee, P.-H. Ren, N. Cai, D. Okamoto, H. W. Choi, and B. Alonso also is acknowledged. This project was supported by U.S. Department of Agriculture National Research Initiative #93-37500-9586, Coors Brewing Co., University of California Agricultural Experiment Station, Cooperative Extension, and BioSTAR Program, Applied Phytologics, Inc., and Novartis, Inc.
Abbreviations
- wtrxh
wheat thioredoxin h gene
- nos
Agrobacterium tumefaciens nopaline synthase 3′ polyadenylation signal
- WTRXh
wheat thioredoxin h protein
- MDH
malate dehydrogenase
- NTR
NADP-thioredoxin reductase
References
- 1.Buchanan B B. Arch Biochem Biophys. 1991;288:1–9. doi: 10.1016/0003-9861(91)90157-e. [DOI] [PubMed] [Google Scholar]
- 2.Jacquot J-P, Lancelin J-M, Meyer Y. New Phytol. 1997;136:543–570. doi: 10.1046/j.1469-8137.1997.00784.x. [DOI] [PubMed] [Google Scholar]
- 3.Besse I, Buchanan B B. Bot Bull Acad Sin (Taipei) 1997;38:1–11. [Google Scholar]
- 4.Kobrehel K, Wong J H, Balogh A, Kiss F, Yee B C, Buchanan B B. Plant Physiol. 1992;99:919–924. doi: 10.1104/pp.99.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lozano R M, Wong J H, Yee B C, Peters A, Kobrehel K, Buchanan B B. Planta. 1996;200:100–106. [Google Scholar]
- 6.Jiao J, Yee B C, Wong J H, Kobrehel K, Buchanan B B. Plant Physiol Biochem. 1993;31:799–804. [Google Scholar]
- 7.Besse I, Wong J H, Kobrehel K, Buchanan B B. Proc Natl Acad Sci USA. 1996;93:3169–3175. doi: 10.1073/pnas.93.8.3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong J H, Jiao J-A, Kobrehel K, Buchanan B B. Plant Physiol. 1995;108:67. (abstr.). [Google Scholar]
- 9.Gautier M-F, Lullien-Pellerin V, De Lamotte-Guery F, Guirao A, Joudrier P. Eur J Biochem. 1998;252:314–324. doi: 10.1046/j.1432-1327.1998.2520314.x. [DOI] [PubMed] [Google Scholar]
- 10.Brandt A, Montembault A, Cameron-Mills V, Rasmussen S K. Carlsberg Res Commun. 1985;50:333–345. [Google Scholar]
- 11.Wan Y, Lemaux P G. Plant Physiol. 1994;104:37–48. doi: 10.1104/pp.104.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemaux P G, Cho M-J, Louwerse J, Williams R, Wan Y. Bio-Rad Bull. 1996;2007:1–6. [Google Scholar]
- 13.Cho M-J, Jiang W, Lemaux P G. Plant Sci. 1998;138:229–244. [Google Scholar]
- 14.Christensen A H, Quail P H. Transgenic Res. 1996;5:1–6. doi: 10.1007/BF01969712. [DOI] [PubMed] [Google Scholar]
- 15.Hunter C P. Ph.D. thesis. University of London, Ashford, Kent: Wye College; 1988. [Google Scholar]
- 16.Choi, H. W., Lemaux, P. G. & Cho, M.-J. (2000) Crop Sci., in press.
- 17.Cho M-J, Choi H W, Buchanan B B, Lemaux P G. Theor Appl Genet. 1999;98:1253–1262. [Google Scholar]
- 18.Dellaporta S. In: in Maize Handbook. Freeling M, Walbot V, editors. New York: Springer; 1993. pp. 522–525. [Google Scholar]
- 19.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Johnson T C, Wada K, Buchanan B B, Holmgren A. Plant Physiol. 1987;85:446–451. doi: 10.1104/pp.85.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 22.Florencio F J, Yee B C, Johnson T C, Buchanan B B. Arch Biochem Biophys. 1988;266:496–507. doi: 10.1016/0003-9861(88)90282-2. [DOI] [PubMed] [Google Scholar]
- 23.Johnson T C, Cao R Q, Kung J E, Buchanan B B, Holmgren A. Planta. 1987;171:321–331. [PubMed] [Google Scholar]
- 24.Serre L, Lauriere C. Anal Biochem. 1990;186:312–315. doi: 10.1016/0003-2697(90)90086-o. [DOI] [PubMed] [Google Scholar]
- 25.Furegon L, Curioni A, Peruffo A D B. Anal Biochem. 1994;221:200–201. doi: 10.1006/abio.1994.1397. [DOI] [PubMed] [Google Scholar]
- 26.Macri L J, MacGregor A W, Schroeder S W. J Cereal Sci. 1993;18:103–106. [Google Scholar]
- 27.MacGregor A W, Macri L J, Schroeder S W. J Cereal Sci. 1994;20:33–41. [Google Scholar]
- 28.Mark D F, Richardson C C. Proc Natl Acad Sci USA. 1976;73:780–784. doi: 10.1073/pnas.73.3.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hardie D G. Phytochemistry. 1975;14:1719–1722. [Google Scholar]
- 30.Sissons M J, Lance R C M. J Cereal Sci. 1993;17:19–24. [Google Scholar]
- 31.Sissons M J, Lance R C M, Wallace W. Cereal Chem. 1994;71:520–521. [Google Scholar]
- 32.Mouaheb N, Thomas D, Verdoucq L, Monfort P, Meyer Y. Proc Natl Acad Sci USA. 1998;95:3312–3317. doi: 10.1073/pnas.95.6.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]