Short abstract
The human malaria parasite Plasmodium falciparum, has many sequences and genes that play key roles in pathogenesis and immune evasion. We must understand the functions of these elements if the chronicity and unpredictable virulence of Plasmodium is to be explained.
Abstract
The human malaria parasite Plasmodium falciparum, one of the world's most devastating pathogens, has an astonishing array of sequences and genes that play key roles in pathogenesis and immune evasion. We must understand the functions of these elements if the chronicity and unpredictable virulence of Plasmodium is to be explained.
Despite intensive efforts over the last century to understand and control malaria, the causative agent of the most severe form of the disease - Plasmodium falciparum - remains firmly entrenched as a leading cause of morbidity and mortality in humans. Approximately 300-500 million clinical episodes and 2.7 million deaths are attributed to P. falciparum infections each year and, with the emergence of widespread drug-resistant parasite populations and insecticide-resistant mosquitoes, this situation is predicted to worsen [1]. New cost-effective strategies for controlling malaria, such as the development of a vaccine, are urgently required. The complete sequence of the 14 linear chromosomes that comprise the P. falciparum genome has recently been determined [2,3,4,5]. It is perhaps not surprising for such a successful pathogen that these studies have revealed that a high proportion of the 5,300 predicted genes encode proteins known or predicted to play a role in pathogenic processes, such as invasion of red blood cells, cytoadherence and immune evasion. We have reviewed elsewhere the impact of the genome sequence on red blood cell invasion [6]; in this article, we comment on our increased understanding of the virulence genes that encode proteins involved in cytoadherence and immune evasion. Insight into the function, diversity and regulation of these genes promises to reveal new strategies for fighting malarial disease [7].
Cytoadherence and antigenic variation
The adhesion of parasite-infected red blood cells to vascular endothelium leads to sequestration of P. falciparum in the deep microvasculature of various tissues and organs, and is associated with certain severe disease outcomes (reviewed in [8,9,10]). A parasite protein inserted into the infected red blood cell surface, known as P. falciparum erythrocyte membrane protein 1 (PfEMP1), is considered to be a key adhesive ligand mediating sequestration. In a process known as antigenic variation, clonal P. falciparum parasites can vary the type of PfEMP1 molecule they express, so as to avoid antibody-mediated clearance. Intriguingly, different PfEMP1 ligands mediate adherence to different receptors on endothelial cells, including the scavenger receptor CD36, chondroitin sulfate A and intracellular adhesion molecule-1. In some instances, parasite populations with a predisposition to adhere to certain receptors are more commonly associated with certain disease outcomes, such as cerebral and placental malaria, although the precise role of parasite-receptor interactions in determining disease severity remains to be understood. P. falciparum infections are persistent, and this chronicity is promoted by antigenic variation at the infected red blood cell surface. Proteins of the repetitive interspersed family (rifins) are also expressed at the surface of infected red blood cells, and, like PfEMP1, these undergo antigenic variation [11,12]. Another family of proteins related to the rifins has also been described - the subtelomeric variable open reading frame (stevor) proteins - but their function remains unknown [11]. PfEMP1 and rifin proteins are considered key virulence factors in P. falciparum.
The PfEMP1, rifin and stevor proteins are encoded by members of the var, rif and stevor gene families, respectively [11,13,14]. As a measure of the significance of these genes to parasite survival, it is now evident that around 10% of the 2.3 megabase P. falciparum genome is committed to the expression and generation of diversity of these virulence genes. This includes the genes themselves - a total of 59 var, 149 rif and 28 stevor genes - as well as intergenic (regulatory) regions and non-coding subtelomeric repeat regions thought to contribute to diversity and transcriptional control of the neighboring virulence genes (these are discussed in more detail below). Members of the var, rif and stevor gene families are mostly concentrated at the end of each P. falciparum chromosome and are positioned directly adjacent to the non-coding subtelomeric repeats (Figure 1); but 23 of the 59 var genes are found in central locations on chromosomes 4, 6, 7, 8 and 12, mostly in head-to-tail arrays of between three and seven genes (see Figure 1, top).
Figure 1.
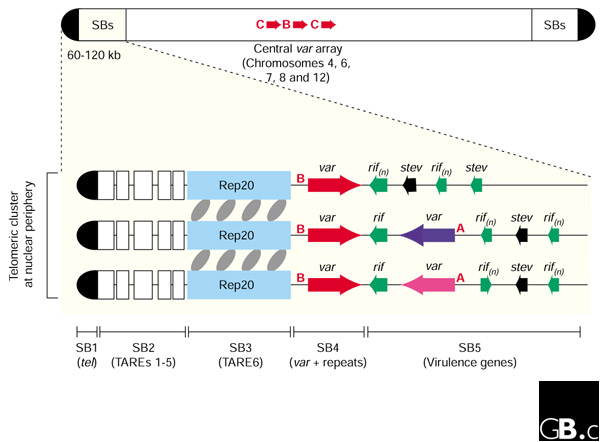
Conserved structures at the ends of P. falciparum chromosomes. A typical chromosome is shown at the top, highlighting the position of subtelomeric blocks (SBs) and centrally located var gene arrays that occur on only a subset of chromosomes. Three typical chromosome end structures are expanded in the lower section and are represented as clustered at the nuclear periphery [19,20]. For all chromosomes, SBs 1-3 are highly conserved in both order and sequence - the individual boxes represent telomere-associated repeat elements (TAREs) 1-6, as previously described [16] - while cross-linking mediated by SB3 (made up of the degenerate 21 base-pair repeat known as Rep20; shown in blue) has been implicated in the stabilization/formation of chromosomal clusters within the cell nucleus (gray ovals) [18]. The first transcribed var gene is typically transcribed toward the centromere in SB4 (for 22 of the 24 chromosomal ends that have a terminal var). The predominant 'type 1' var genes (colored red) are found in the positions shown. A, B and C represent var upstream sequences, upsA, upsB and upsC, respectively, that are typically found upstream of var genes in the locations shown (note that upsB is found upstream of both subtelomeric and internal var genes). The rif, stevor and var genes in SB5 have a common arrangement in some, but not all, chromosome ends. For example, the arrangement of telomere-var-rif/stevor(n) shown at the top of the cluster in the figure is found at 10 chromosome ends, but even among this group there is variation in the arrangement and orientation of transcription of the rif and stevor genes. Hence, although chromosomal clustering within the nucleus promotes ectopic recombination and the generation of diversity in virulence genes [15,19], this is likely to be limited by the variation in the gene structure within SB5.
The subtelomeric region
From the genome sequence, it is clear that most of the 28 P. falciparum chromosome ends are structurally highly conserved, and each end can be divided into five different subtelomeric blocks comprising up to 120 kilobases each (Figure 1) [2]. Three subtelomeric blocks (SB1-SB3) are non-coding, while the remaining two (SB4 and SB5) incorporate virulence-gene family members amongst other sequences (Figure 1). SB1 is located at the extreme terminus and is comprised of approximately 1.2 kb of a seven base-pair G-rich telomere repeat with the consensus sequence GGGTT(T/C)A [15]. SB2 and SB3 together are 30-40 kb in length and comprise six different non-coding repeat elements previously known as telomere-associated repeat elements (TAREs) 1-6 [15,16].
Although subtelomeric repeats have been identified in other organisms, the P. falciparum SB2 and SB3 elements are unique in eukaryotes, and even amongst the Plasmodia, in terms of their size, composition and complexity. SB2 comprises a series of five ordered repeat elements (TAREs 1-5) that are interspersed with non-repetitive sequence. These elements are always found in the same order, and they display remarkable sequence conservation and a CG-bias (around 30% G+C) that is unusual in the non-coding regions of the otherwise extremely AT-rich genome (normally about 10% G+C in non-coding regions and 19.4% G+C overall). It is possible that SB2 (TAREs 1-5) represents a distinct functional unit, although this remains to be examined. SB3 (or TARE6) consists entirely of a 10-20 kb stretch of a unique degenerate 21 base-pair repeat known as Rep20 [17,18]. More repeat elements are found in SB4, but this region also contains at least one var gene. Hence, the most telomeric var genes, which are usually transcribed in a telomere-to-centromere direction, are the first genes transcribed at most chromosome ends (in total, in 24 of 28 ends). In some chromosome ends, fragments of SB4 are inverted. SB5 may extend up to 120 kb inwards, towards the centromere, and contains members of the var, rif, stevor and other gene families.
The conserved arrangement of P. falciparum chromosome ends is thought to mediate chromosome-end alignment and clustering in a manner that promotes recombination in telomere-associated genes located at the ends of heterologous chromosomes (ectopic recombination). This has been demonstrated to occur in var genes, resulting in gene conversion events [19]. The process of ectopic recombination in var genes, which is also likely to occur in other subtelomeric gene family members, allows the rapid generation of diverse antigenic and adherence phenotypes. Recent evidence suggests that elements in SBs 2-5, and in particular SB3, are necessary for chromosome-end clustering but not for the anchoring of chromosome ends at the nuclear periphery, a phenomenon that is probably mediated by SB1 (the telomere tract) [18,20]. Hence, parasite populations and individual clonal lines have distinct complements of virulence genes. It is interesting to note that the complement of var genes in the sequenced P. falciparum genome is dominated by one particular var type (var types are classified according to their particular arrangement of encoded cysteine-rich domains). Although 16 different var types were identified in the sequenced 3D7 genome, 38 of the 59 var genes in this parasite line were of the same four-domain type, termed 'type 1' [2]. It is striking that the 38 type 1 var genes are not distributed randomly but are found either at the extreme telomere, where they are always present in a telomere-to-centromere orientation, or in central chromosomal locations, where they comprise 20 of the 23 var genes in this region. The dominance of one var type was unexpected, as previously characterized var genes to which an adhesive phenotype had been assigned were generally not of this type, and indeed encoded more than four domains.
The mechanisms that control expression of the P. falciparum virulence genes remain poorly understood. In the case of var genes, at any one time all but one are repressed. A cooperative interaction - which does not depend on chromosomal context - between the var promoter and intron regions can mediate this silencing [21], but the mechanism(s) that control var gene activation and switching remain to be determined. It is interesting that the upstream regions of the 59 var genes in the sequenced genome could be classified into three distinct classes, termed upsA, upsB and upsC [2]. These regions are not distributed randomly but are generally associated with particular var genes: upsA with subtelomeric genes transcribed toward the telomere; upsB with subtelomeric genes transcribed toward the centromere; and upsC with 13 centrally located var genes; the remaining 10 internal var genes have the upsB-type 5'-untranslated region. The upsB and upsC are likely to be the same as the two var promoter elements that have previously been identified [22]; but the functional relevance of these three different upstream region types remains unknown.
Beyond the genome
The genome sequence of P. falciparum has revealed a long-suspected but nevertheless breathtaking array of sequences and genes known or suspected to mediate virulence. Much remains to be learned about the mechanisms that control the expression, switching, and generation of allelic diversity of P. falciparum virulence genes, as well as about the nature of the adhesive and antigenic phenotypes of the proteins encoded by these genes. As a result of the genome project, such studies are now possible, and these are likely to provide fertile ground for researchers in the coming decade. It is hoped that such understanding will have significant impact on control measures that aim to alleviate the misery of malaria.
Acknowledgments
Acknowledgements
We thank Rebecca O'Donnell and Till Voss for critical reading of this manuscript. B.S.C. and A.F.C. are International Research Fellows of the Howard Hughes Medical Institute.
Contributor Information
Brendan S Crabb, Email: crabb@wehi.edu.au.
Alan F Cowman, Email: cowman@wehi.edu.au.
References
- Greenwood B, Mutabingwa T. Malaria in 2002. Nature. 2002;415:670–672. doi: 10.1038/415670a. [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Nelson KE, Bowman S, Paulsen IT, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MJ, Shallom SJ, Carlton JM, Salzberg SL, Nene V, Shoaibi A, Ciecko A, Lynn J, Rizzo M, Weaver B, et al. Sequence of Plasmodium falciparum chromosomes 2, 10, 11 and 14. Nature. 2002;419:531–534. doi: 10.1038/nature01094. [DOI] [PubMed] [Google Scholar]
- Hall N, Pain A, Berriman M, Churcher C, Harris B, Harris D, Mungall K, Bowman S, Atkin R, Baker S, et al. Sequence of Plasmodium falciparum chromosomes 1, 3-9 and 13. Nature. 2002;419:527–531. doi: 10.1038/nature01095. [DOI] [PubMed] [Google Scholar]
- Hyman W, Fung E, Conway A, Kurdi O, Mao J, Miranda M, Nakao B, Rowley D, Tamaki T, Wang F, et al. Sequence of Plasmodium falciparum chromosome 12. Nature. 2002;419:534–537. doi: 10.1038/nature01102. [DOI] [PubMed] [Google Scholar]
- Cowman AF, Crabb BS. The Plasmodium falciparum genome - a blueprint for erythrocyte invasion. Science. 2002;298:126–128. doi: 10.1126/science.1078169. [DOI] [PubMed] [Google Scholar]
- Baruch DI, Gamain B, Barnwell JW, Sullivan JS, Stowers A, Galland GG, Miller LH, Collins WE. Immunization of Aotus monkeys with a functional domain of the Plasmodium falciparum variant antigen induces protection against a lethal parasite line. Proc Natl Acad Sci USA. 2002;99:3860–3865. doi: 10.1073/pnas.022018399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeson J, Reeder J, Rogerson S, Brown G. Parasite adhesion and immune evasion in placental malaria. Trends Parasitol. 2001;17:331–337. doi: 10.1016/S1471-4922(01)01917-1. [DOI] [PubMed] [Google Scholar]
- Craig A, Scherf A. Molecules on the surface of the Plasmodium falciparum infected erythrocyte and their role in malaria pathogenesis and immune evasion. Mol Biochem Parasitol. 2001;115:129–143. doi: 10.1016/S0166-6851(01)00275-4. [DOI] [PubMed] [Google Scholar]
- Kyes S, Horrocks P, Newbold C. Antigenic variation at the infected red cell surface in malaria. Annu Rev Microbiol. 2001;55:673–707. doi: 10.1146/annurev.micro.55.1.673. [DOI] [PubMed] [Google Scholar]
- Cheng Q, Cloonan N, Fischer K, Thompson J, Waine G, Lanzer M, Saul A. stevor and rif are Plasmodium falciparum multicopy gene families which potentially encode variant antigens. Mol Biochem Parasitol. 1998;97:161–176. doi: 10.1016/S0166-6851(98)00144-3. [DOI] [PubMed] [Google Scholar]
- Kyes SA, Rowe JA, Kriek N, Newbold CI. Rifins: a second family of clonally variant proteins expressed on the surface of red cells infected with Plasmodium falciparum. Proc Natl Acad Sci USA. 1999;96:9333–9338. doi: 10.1073/pnas.96.16.9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, Taraschi TF, Howard RJ. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- Su XZ, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, Ravetch JA, Wellems TE. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- Scherf A, Figueiredo LM, Freitas-Junior LH. Plasmodium telomeres: a pathogen's perspective. Curr Opin Microbiol. 2001;4:409–414. doi: 10.1016/S1369-5274(00)00227-7. [DOI] [PubMed] [Google Scholar]
- Figueiredo LM, Pirrit LA, Scherf A. Genomic organisation and chromatin structure of Plasmodium falciparum chromosome ends. Mol Biochem Parasitol. 2000;106:169–174. doi: 10.1016/S0166-6851(99)00199-1. [DOI] [PubMed] [Google Scholar]
- Aslund L, Franzen L, Westin G, Persson T, Wigzell H, Pettersson U. Highly reiterated non-coding sequence in the genome of Plasmodium falciparum is composed of 21 base-pair tandem repeats. J Mol Biol. 1985;185:509–516. doi: 10.1016/0022-2836(85)90067-1. [DOI] [PubMed] [Google Scholar]
- O'Donnell RA, Freitas-Junior LH, Preiser PR, Williamson DH, Duraisingh M, McElwain TF, Scherf A, Cowman AF, Crabb BS. A genetic screen for improved plasmid segregation reveals a role for Rep20 in the interaction of Plasmodium falciparum chromosomes. EMBO J. 2002;21:1231–1239. doi: 10.1093/emboj/21.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas-Junior LH, Bottius E, Pirrit LA, Deitsch KW, Scheidig C, Guinet F, Nehrbass U, Wellems TE, Scherf A. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- Figueiredo LM, Freitas-Junior LH, Bottius E, Olivo-Marin JC, Scherf A. A central role for Plasmodium falciparum subtelomeric regions in spatial positioning and telomere length regulation. EMBO J. 2002;21:815–824. doi: 10.1093/emboj/21.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitsch KW, Calderwood MS, Wellems TE. Malaria. Cooperative silencing elements in var genes. Nature. 2001;412:875–876. doi: 10.1038/35091146. [DOI] [PubMed] [Google Scholar]
- Voss T, Thompson J, Waterkeyn J, Felger I, Weiss N, Cowman A, Beck H. Genomic distribution and functional characterisation of two distinct and conserved Plasmodium falciparum var gene 5' flanking sequences. Mol Biochem Parasitol. 2000;107:103–115. doi: 10.1016/S0166-6851(00)00176-6. [DOI] [PubMed] [Google Scholar]


