Abstract
One of the challenges faced in malarial control is the acquisition of insecticide resistance that has developed in mosquitoes that are vectors for this disease. Anopheles gambiae, which has been the major mosquito vector of the malaria parasite Plasmodium falciparum in Africa, has over the years developed resistance to insecticides including dieldrin, 1,1-bis(p-chlorophenyl)-2,2,2-trichloroethane (DDT), and pyrethroids. Previous microarray studies using fragments of 230 An. gambiae genes identified five P450 loci, including CYP4C27, CYP4H15, CYP6Z1, CYP6Z2, and CYP12F1, that showed significantly higher expression in the DDT-resistant ZAN/U strain compared with the DDT-susceptible Kisumu strain. To predict whether either of the CYP6Z1 and CYP6Z2 proteins might potentially metabolize DDT, we generated and compared molecular models of these two proteins with and without DDT docked in their catalytic sites. This comparison indicated that, although these two CYP6Z proteins share high sequence identity, their metabolic profiles were likely to differ dramatically from the larger catalytic site of CYP6Z1, potentially involved in DDT metabolism, and the more constrained catalytic site of CYP6Z2, not likely to metabolize DDT. Heterologous expressions of these proteins have corroborated these predictions: only CYP6Z1 is capable of metabolizing DDT. Overlays of these models indicate that slight differences in the backbone of SRS1 and variations of side chains in SRS2 and SRS4 account for the significant differences in their catalytic site volumes and DDT-metabolic capacities. These data identify CYP6Z1 as one important target for inhibitor design aimed at inactivating insecticide-metabolizing P450s in natural populations of this malarial mosquito.
Keywords: cytochrome P450 monooxygenases, insecticides, plant allelochemicals
In 2004, the World Health Organization reported that up to 2.7 million people die of malaria every year with 80–90% of these deaths occurring in Africa (ref. 1 and www.africanfront.com/AIDS1.php). Many prevention and treatment strategies have been developed to tackle this life-threatening disease from the side of the mosquito vector and that of the human host (2, 3). These range from antimalarial drugs to indoor spraying of insecticides and use of bed nets treated with pyrethroid insecticides. Although these practices have helped reduce human mortality, various issues have emerged with mosquito vectors developing insecticide resistance and parasites developing drug resistance. Both of these significantly reduce the efficacy of current malarial prevention and treatment practices (2, 4–8).
The development of insecticide resistance in mosquito vectors is illustrated by Anopheles gambiae, which has been the major mosquito vector of the malaria parasite Plasmodium falciparum in Africa (4, 6). Throughout the years, people have reported Anopheles resistance (albeit low-level resistance) to various insecticides including dieldrin (a cyclodiene-type insecticide), 1,1-bis(p-chlorophenyl)-2,2,2-trichloroethane (DDT), and permethrin (a pyrethroid-type insecticide) (9). The completed genome sequence of An. gambiae that became available in 2002 (10) opened up the possibility of understanding the molecular basis of insecticide resistance in this species. The initial identification of genes potentially responsible for resistance to permethrin and DDT analyzed variations in the transcript levels of 230 An. gambiae genes on microarrays containing gene-specific probes (11). Represented on the array were members of several large gene families likely to be associated with insecticide resistance including cytochrome P450 monooxygenases (P450s), GSTs, carboxyl/cholinesterases, redox genes, and ABC transporters as well as genes for tissue-specific and housekeeping functions. The range of genes showing >1.5-fold overexpression in the DDT-resistant strain (ZAN/U) compared with the susceptible strain (Kisumu) included five P450 loci (CYP4C27, CYP4H15, CYP6Z1, CYP6Z2, and CYP12F1), one GST locus (GSTE2), and four peroxidase loci (PX9, PX12, PX13A, and PX13B) (11). The range of genes showing >1.5-fold underexpression in this DDT-resistant strain compared with the susceptible strain included four P450s (CYP6M4, CYP6AK1, CYP9J5, and CYP12F2), GSTT2, an esterase (COE13O), a thioredoxin reductase (TRX2), and a thioredoxin peroxidase (TPX4). These significant variations in transcript accumulation between the DDT-resistant and -susceptible strains suggested that one or several of these proteins may have roles in DDT detoxification. Although higher expression in insecticide-resistant strains does not necessarily guarantee relevance to insecticide resistance (12), CYP6Z1 has been repeatedly highlighted as overexpressed in adult males and females of the permethrin-resistant RSP strain collected in West Kenya (East Africa) and constitutively expressed at low levels in larvae and pupae of the RSP strain (13, 14). And CYP6Z2 (but not CYP6Z1) has been documented as overexpressed in adult males and females of the permethrin-resistant Odumasy strain collected in Southern Ghana (West Africa) (15).
Noting that DDT-metabolizing and cypermethrin-metabolizing P450s have already been identified in the CYP6 family of dipterans (Drosophila melanogaster and Musca domestica) and lepidopterans (Helicoverpa zea) (16–22), the overall purpose of this study was to determine whether any of CYP6 family transcripts expressed at higher levels in DDT-resistant strains have potential to code for a P450 capable of metabolizing DDT. Because we were specifically interested in comparing the metabolic profiles of the closely related CYP6Z1 and CYP6Z2 (69.6% amino acid identity), molecular modelings were carried out to predict their catalytic site geometries. These comparisons indicated that, although the two proteins share high primary sequence identity, their predicted substrate cavities are dramatically different, with CYP6Z1 predicted to metabolize DDT and CYP6Z2 predicted not to bind this insecticide. Subsequent biochemical characterizations supported these predictions and identified CYP6Z1 as a potential target for the design of inhibitors capable of reducing An. gambiae resistance to DDT and other insecticides.
Results
Development of the CYP6Z Models.
As mentioned, our predictive molecular modeling exercise was originally aimed at comparing the structures of An. gambiae CYP6Z1 and CYP6Z2 to describe their potential for metabolizing DDT. The first step in the model-building process was to select and align a P450 template chosen from available eukaryotic P450 crystal structures with the CYP6Z sequences maximizing sequence identities and functional residues (23–25). To ensure that the differences in the CYP6Z1 and CYP6Z2 models were not the consequence of differences in the alignment of the two sequences with the template structure, the CYP6Z1 and CYP6Z2 sequences from the RSP-ST and PEST strains, respectively, were first aligned to one another by using the ALIGN function in the MOE program that showed 69.6% sequence identity between the two proteins (Fig. 1). This pairwise alignment was then fixed and subsequently aligned with a fixed alignment of eight class II (eukaryotic) P450s as in Baudry et al. (24). Of these, human CYP3A4 (Protein Data Bank ID code 1TQN) (26) shared the highest sequence identity with CYP6Z1 (28.1%) and CYP6Z2 (28.7%), and it was used with the CYP6Z pair set to create a final three-sequence alignment.
Fig. 1.
Sequence alignments of CYP6Z1, CYP6Z2, and CYP3A4. An. gambiae CYP6Z1 from the RSP-ST strain (GenBank accession no. AF487535), An. gambiae CYP6Z2 from the PEST strain (GenBank accession no. XP_317252), and human CYP3A4 (Protein Data Bank ID code 1TQN) are shown with underlines representing α-helices and gray blocks representing SRS regions. Asterisks represent identical residues, colons represent conserved residues, and dots represent semiconserved residues in the three sequences. Residues in the CYP6Z1 sequence predicted to exist within 4.5 Å of DDT are boxed.
Evaluation of the Computer Models of CYP6Z1 and CYP6Z2 Models.
The molecular models of CYP6Z1 and CYP6Z2 were generated as described in Materials and Methods, and the final substrate-free models were examined for the distribution of ϕ and ψ angles by using Ramachandran plots generated within MOE. This evaluation indicated that only 11 of 475 residues (2.3%) in the CYP6Z1 model and 12 of 474 residues (2.5%) in the CYP6Z2 model were outliers. Because all of these residues were located in the exterior regions of the models, no attempts were made to further refine the models. Prosa II analysis applied to test the native-fold likeness of these models indicated that both CYP6Z1 and CYP6Z2 have low positive Prosa II scores across most of their length [supporting information (SI) Fig. S1], indicating that they are correctly folded. These and other evaluations outlined in Materials and Methods strongly supported the quality of both models. Even so, it must be acknowledged that the necessity of using low-identity P450 templates causes some inherent uncertainty in the process of predictive modeling, especially in the three most variable loops outlined by Baudry et al. (24).
Structural Analysis of the CYP6Z1 and CYP6Z2 Models.
Because a common template (CYP3A4) was used, the backbone conformations of the CYP6Z1 and CYP6Z2 models overlaid in Fig. 2 are within 2.0 Å rmsd except for 22 residues located in the following regions: the N terminus of the A′–A loop, the N terminus of the loop between helix A and β1-1 sheet, the C terminus of the F′ helix, the N terminus of the F′–G′ loop, the C terminus of the H helix, the N terminal half of the H–I loop, the middle of the J–K loop, and the N terminus of the loop between the β2-1 and β2-2 sheets. Substrate recognition sites (SRS) in CYP6Z1 and CYP6Z2 were identified by aligning with H. zea CYP6B8v1 (27), which shares higher identity with these mosquito P450s (29.6% to CYP6Z1 and 30.3% to CYP6Z2) than any other class II P450 (13–24% sequence identity). From the perspective of tertiary structure, SRS1 corresponds to the loop region between the B and C helices positioned over the heme, SRS2 and SRS3 correspond to the F and G helices that make up part of the substrate access channel, SRS4 corresponds to the long I helix extending through the “back” of the catalytic site, and SRS5 and SRS6 correspond to the N terminus of β-strand 1–4 and the β-turn at the end of β-sheet 4, respectively, which both protrude into the catalytic site. From the perspective of primary sequence, SRS1–SRS6 span residues 97–121, 207–213, 233–240, 287–305, 362–371, and 473–480 as highlighted in the CYP6Z1 and CYP6Z2 sequences shown in Fig. 1. The number of conserved residues in SRS1–SRS6 are 22/25, 2/7, 3/8, 16/19, 7/10, and 2/8, indicating that SRS1, SRS4, and SRS5 are more conserved than SRS2, SRS3, and SRS6. The predicted SRS backbones of these six regions are overlaid in Fig. 2 with individual SRS regions shown in Fig. 3. All backbone conformations in these SRS regions are within 2.0 Å rmsd except for the loop between SRS2 and SRS3 and a few other regions located outside of these SRS regions.
Fig. 2.
Backbone overlays of the CYP6Z1 and CYP6Z2 models. Backbone overlays of the CYP6Z1 and CYP6Z2 models are shown with the SRS1–SRS6 regions shown in purple and the heme in the floor of the catalytic site shown in elemental colors.
Fig. 3.
Conserved and variant side chains within the SRS regions of CYP6Z1 and CYP6Z2. Backbone and side chain overlays in the six SRS regions of the CYP6Z1 and CYP6Z2 substrate-free models are shown with conserved side chains in light gray and variant side chains in black. CYP6Z1 residues are depicted in ball-and-stick format, and CYP6Z2 residues are depicted in stick format. (a) SRS1. (b) SRS2 and SRS3. (c) SRS4 and SRS5. (d) SRS6.
To identify the possible substrate access channels for the substrate-free CYP6Z1 and CYP6Z2 models, all hydrophobic and hydrophilic cavities inside of these models were searched separately by using the ALPHA SITE FINDER function within MOE (28). Two possible substrate access channels that form continuous pathways from the substrate binding pocket to the protein surface exist in the CYP6Z1 model. These are designated pw2a and pw3 following the substrate access channel nomenclature of Wade et al. (29). The pw2a channel extends from the heme to the exterior of the protein through an opening encircled by the F and G helices, the B′ helix and the β1-1 sheet, and the pw3 channel extends between the F and G helices.
Docking of Prospective Substrates into the CYP6Z1 and CYP6Z2 Models.
To predict whether DDT and/or carbaryl might be substrates for CYP6Z1 and CYP6Z2, these insecticides were docked within each predicted catalytic site by using DOCK function within MOE after attaching a singlet oxygen to the heme plane using parameter assignments specified by Rupasinghe et al. (30). After their initial placement above the heme plane, docking simulations were run by using the MMFF94s force field (31–36) and the simulated annealing conformation search method of the DOCK function. In the initial runs, 100–300 potential conformations for DDT and 100 potential conformations for both carbaryl and xanthotoxin were generated while keeping the protein side chains fixed in their orientations. The docked conformations were sorted in ascending order with respect to the total energy (the sum of van der Waals as well as electrostatic energy terms of the potential energy function and the ligand's internal energy). The binding modes with the lowest total energies and known hydroxylation sites [trichloromethyl group in DDT (20, 37); carbons 4–7 in carbaryl (38); furan ring in xanthotoxin (39)] closest to the heme were chosen for the second round of energy minimization during which all side chains were allowed to move freely.
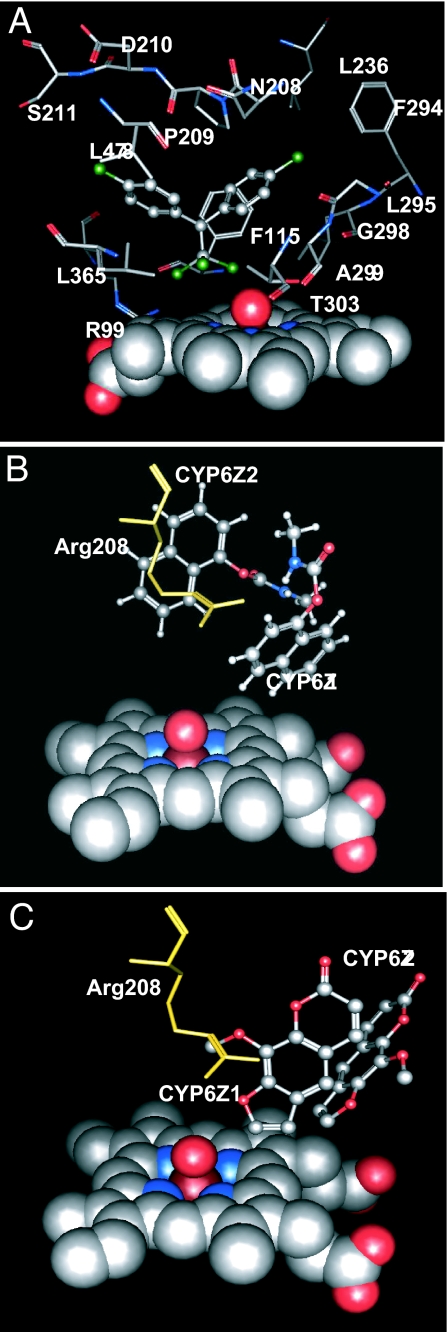
The predicted conformation of DDT in the CYP6Z1 model (Fig. 4A) shows that this compound bound with its trichloromethyl group positioned at a distance of 4.57 Å from the oxygen of the Fe-O catalytic intermediate with an interaction energy of −39.5 kcal/mol after two rounds of energy minimization. In this binding mode, the following residues are within 4.5 Å of DDT: R99 and F115 in SRS1; N208, P209, D210, and S211 in SRS2; L236 in SRS3; F294, L295, G298, A299, and T303 in SRS4; L365 in SRS5; and L478 in SRS6 (all are boxed in Fig. 1). Although CYP6Z2 was subjected to several rounds of docking simulations to ensure thorough searching for a DDT binding mode, none of the top-ranked docked-DDT conformations positioned the trichloromethyl group in any reasonable proximity to the oxygen of the Fe-O intermediate (Table S1 and Fig. S2), suggesting that CYP6Z2 might not metabolize this chlorinated insecticide.
Fig. 4.
Predicted substrate binding modes in the CYP6Z1 and CYP6Z2 models. (A) The lowest-energy binding mode for DDT (ball-and-stick format) in the CYP6Z1 model is shown with residues within 4.5 Å of this substrate (stick format) and the heme-oxo complex (space-filling format). (B) The lowest-energy binding modes for carbaryl (ball-and-stick format) in the CYP6Z1 and CYP6Z2 models are shown with Arg-208 of CYP6Z2 (stick format) extending into the catalytic site. (C) The lowest-energy binding modes for xanthotoxin (ball-and-stick format) in the CYP6Z1 and CYP6Z2 models are shown with Arg-208 of CYP6Z2 (stick format) extending into the catalytic site.
In comparison, the smaller carbaryl molecule docked in the CYP6Z1 model with carbons 4 and 5 positioned at distances of 4.4 and 3.7 Å, respectively, from the heme oxygen (Fig. 4B) with an interaction energy of −42.0 kcal/mol. Carbaryl could also be accommodated in the CYP6Z2 model with carbons 6 and 7 positioned at ≈5.5 Å from the heme oxygen with an interaction energy of −33.0 kcal/mol.
To determine whether toxic allelochemicals potentially encountered in plant litter might also be metabolized, the planar furanocoumarin xanthotoxin was docked in both models. It is predicted to be positioned with its furan ring at a distance of 2.6 Å from the heme in the CYP6Z1 model and at the significantly greater distance of 6.7 Å in the CYP6Z2 model (Fig. 4C).
Comparative Metabolism Assays.
With the inevitable uncertainties of molecular models, the activities of CYP6Z1 and CYP6Z2 were compared in side-by-side activity assays of these proteins coexpressed with housefly P450 reductase (HFR) and fruit fly cytochrome b5 (b5) at a multiplicity of infection ratio of 2:2:0.1 (P450:HFR:b5). These assays indicated that both proteins metabolize carbaryl with CYP6Z1 functioning 4.5-fold faster than CYP6Z2 (Table 1). CYP6Z1 also metabolizes DDT at a significant rate (3.91 nmol/min per nanomole of P450) and metabolizes xanthotoxin at a more modest rate (1.51 nmol/min per nanomole of P450), whereas CYP6Z2 does not metabolize either of these chemicals.
Table 1.
Catalytic activities of CYP6Z proteins coexpressed in Sf9 cells with house fly P450 reductase
| Substrate | CYP6Z1 | CYP6Z2 |
|---|---|---|
| DDT | 3.91 (0.67) | ND |
| Carbaryl | 2.20 (0.14) | 0.49 (0.09) |
| Xanthotoxin | 1.51 (0.11) | ND |
Data show metabolic activity (with SE in parentheses) in nanomoles per minute per nanomole of P450.
Binding Site Comparisons.
The dramatic differences in the CYP6Z1 and CYP6Z2 dockings and catalytic activities were further evaluated by detailed analysis of the predicted binding sites by using Computed Atlas of Surface Topography of proteins (CASTp) analysis (40). From this, the binding site volumes without various channels extending to the protein surface were estimated to be 3,471.1 Å3 and 2,601.9 Å3 for CYP6Z1 and CYP6Z2, respectively, indicating that the binding site of CYP6Z2 is considerably smaller than that of CYP6Z1.
Additional overlays of the DDT-docked CYP6Z1 model (green in Fig. 5) with the CYP6Z2 substrate-free model (gold in Fig. 5) predict that van der Waals clashes between the DDT molecule and side chains in the CYP6Z2 catalytic site block binding of this compound. In ball-and-stick representation in Fig. 5, problematic CYP6Z2 residues include Phe-115, Arg-208, and Ile-298 in SRS1, SRS2, and SRS4. With the side chain of Phe-115 in the B–C loop (SRS1) showing the least variation in positioning between the two models, the CYP6Z2 model suggests that SRS1 protrudes farther into the catalytic site than in the CYP6Z1 model causing Phe-115 in CYP6Z2 to have van der Waals clashes with DDT whereas Phe-115 in CYP6Z1 does not. Without displaying van der Waals clashes, the side chain of Arg-208 in CYP6Z2 is pushed substantially farther into the binding site whereas Asn-208 in CYP6Z1 lies within contact distance of DDT. The side chain of Ile-298 in CYP6Z2 is significantly longer than the side chain of Gly-298 in CYP6Z1, and it occurs at a position in the I helix that may have substantial impact on substrate binding.
Fig. 5.
Overlays of the CYP6Z2 substrate-free model on the DDT-docked CYP6Z1 model. The substrate-free CYP6Z2 model overlaid on the DDT-docked CYP6Z1 model is shown with the DDT molecule in gray ball-and-stick format. All side chains within 4.5 Å of DDT docked in CYP6Z1 are shown in green stick format, and those in CYP6Z2 are shown in gold stick format. The two CYP6Z2 side chains that display the most problematic van der Waals contacts with DDT (Arg-208 and Ile-298) are rendered in gold ball-and-stick format. Phe-115 that is identical in these two proteins but displays van der Waals contacts with DDT in the CYP6Z2 model is shown in stick format behind Ile-298.
Sequence Variants.
With the CYP6Z1 model developed from a cDNA sequence available from the permethrin- and DDT-resistant RSP-ST strain (41, 42) and the CYP6Z2 model developed from a cDNA sequence available from the insecticide-susceptible PEST strain, we further evaluated a number of sequence variants deposited in the GenBank database for other Anopheles strains. Alignment of the CYP6Z1 RSP-ST sequence with the CYP6Z1 sequence expressed from the G3 strain identified four differences (S107N, R327Q, T346N, and V492A listed with RSP-ST residue first and G3 residue second). Alignment of the CYP6Z1 RSP-ST sequence with the CYP6Z1 sequence from the PEST strain identified six differences (A11T, M213I, M261T, R327Q, T346N, and V492A listed with RSP-ST residue first and PEST residue second). Comparisons between these three sequences indicate that the expressed CYP6Z1 G3 protein is identical in its first 326 aa to the RSP-ST sequence (except for a single S107N change) and identical in its last 233 aa to the PEST sequence (corresponding to a 59-aa overlap of these three sequences). Of the residues in SRS, M213I is located in SRS2 in proximity to other residues predicted to contact DDT (i.e., N208, P209, D210, and S211), and S107N is located in SRS1 in a portion of the B–C loop predicted to be outside substrate contact range. The other non-SRS residues exist in the signal sequence (A11T), the G–H loop (M261T) and the J helix (R327Q) on the protein surface, the K helix (T346N) on the proximal surface in a region potentially interacting with P450 reductase, and at the extreme C terminus (V492A). To determine whether any of these changes might affect the predicted binding mode for DDT, the CYP6Z1 RSP-ST model was mutated to the CYP6Z1 PEST sequence by using ROTAMER EXPLORER and MUTATE functions within MOE. After selecting the rotamer with the highest-ranking score as the rotamer of each replacement side chain, another energy-minimization run was conducted on the DDT docked model to obtain the CYP6Z1 PEST model. Compared with an interaction energy of −39.5 kcal/mol and distance to the trichloromethyl group on DDT of 4.57 Å for the CYP6Z1 RSP-ST model, the interaction energy and distance to the heme oxygen are −38.63 kcal/mol and 4.54 Å for the CYP6Z1 PEST model. These small differences suggest that the catalytic sites for these more divergent variants are essentially the same. In addition, alignment of the CYP6Z1 PEST genomic sequence (http://p450.antibes.inra.fr) with another reported CYP6Z1 PEST genomic sequence (GenBank accession no. EAA45654) indicated that the second variant in this strain has an unexplained 3-nt deletion of I165 in the loop region between the D and E helices and four amino acid replacements (T11A, V90M, V166S, and A492V) compared with the sequence presented at http://p450.sophia.inra.fr; the first and last of these changes correspond to sequences in the RSP-ST strain that we have already described. Again, none of these changes is predicted to impact DDT binding and metabolism.
Among the available CYP6Z2 variants, alignment of the modeled CYP6Z2 PEST sequence with the expressed CYP6Z2 G3 sequence identified one difference (S210T listed with PEST residue first and G3 residue second) in the F helix outside of substrate contact range. Alignment of the CYP6Z2 PEST sequence with the CYP6Z2 RSP-ST genomic sequence (42) identified another difference (K458N listed with PEST residue first and RSP-ST residue second) in a loop on the protein surface. Another CYP6Z2 cDNA sequence reported from the RSP-ST strain (AF487780) appears to be missing two amino acids (L441 and R442) immediately downstream from the heme cysteine because of alternate processing of the single intron in this transcript. Loss of these two residues is predicted to shorten the L helix beneath the heme and potentially affect hydrogen-bonding interactions with P450 reductase. It is not yet clear whether the six nucleotides responsible for this deletion are missing in other CYP6Z2 transcripts.
Discussion
Given that their x-ray crystal structures are not available, homology models have allowed us to construct and dock models of CYP6Z1 and CYP6Z2 with DDT, a chlorinated insecticide, carbaryl, a carbamate insecticide, and xanthotoxin, a plant toxin. Our results predicted that these two CYP6Z subfamily proteins have dramatically different catalytic sites even though they share 69.6% overall sequence identity. For CYP6Z1, the highest-ranking docking conformation for DDT positions its trichloromethyl group at a distance of 4.57 Å from the heme oxygen bound to the heme iron with an interaction energy of −39.5 kcal/mol. For CYP6Z2, repeated rounds of DDT docking were unable to identify binding modes appropriate for metabolism of DDT. Docking of xanthotoxin in these models predicted that it could bind in close enough proximity to the heme in CYP6Z1 for catalytic turnover, but not in CYP6Z2. In contrast, the docking modes for carbaryl suggested that, although different in their predicted binding orientations, both CYP6Z1 and CYP6Z2 should be capable of binding and metabolizing it in at least one position.
Although there may be some uncertainties in some regions of these CYP6Z models because of their low sequence identity with the template used, comparisons of their predicted catalytic sites provide several explanations for the significant differences in their DDT docking results. The first difference is that, even with backbone conformations overlaying one another in the SRS regions, the binding site volume is much smaller for CYP6Z2 because of long side chain extensions into the catalytic site. As shown in Fig. 4A, almost all residues located within 4.5 Å of DDT in the docked CYP6Z1 model are close in position to their analogous side chains in CYP6Z2; the two exceptions to this (Gly-209 in CYP6Z2/Pro-209 in CYP6Z1 and Arg-210 in CYP6Z2/Asp-210 in CYP6Z1) occur in SRS2 in the upper regions of the catalytic site where changes in side-chain volume can affect positioning of the two aromatic rings on this insecticide. In this region, the side chains of CYP6Z1 are predicted to be distant enough from DDT to be free of van der Waals clashes. Contrasting with this, three residues in CYP6Z2 (Phe-115, Ile-298, and, especially, Arg-208) protrude far enough into the catalytic site that they constrain its volume, causing even top-ranked DDT docking modes to have high interaction energies and inappropriate positionings over the heme oxygen.
Metabolic analyses of heterologously expressed CYP6Z1 and CYP6Z2 from the G3 strain have clearly supported our predictions. CYP6Z1, which is reported to be overexpressed in at least two DDT- and permethrin-resistant strains (11, 13, 14), is capable of metabolizing DDT, carbaryl, and xanthotoxin. CYP6Z2, which is reported to be overexpressed in the same DDT-resistant strain and another permethrin-resistant strain (11, 15), is capable of metabolizing carbaryl but not DDT or xanthotoxin.
The fact that closely related P450 sequences have different metabolic profiles is certainly not new in the P450 field. As reviewed for vertebrate P450s (43), the closely related pairs of CYP2A4 and CYP2A5, CYP2C4 and CYP2C5, and CYP3A4 and CYP3A5 with sequence identities of 98%, >95%, and >85%, respectively, do not metabolize the same substrates with similar rates or regioselectivities. Among plant P450s, spearmint CYP71D15 and peppermint CYP71D18 have different regiospecificities for limonene hydroxylation because of a single amino acid difference within the catalytic site (F363I in SRS5) (44, 45). The closely related lepidopteran CYP6B1 and CYP6B3 also differ in their metabolism of plant allelochemicals (46). These metabolic variations highlight the fact that, even with high global sequence identity, only a small number of residues in each binding site define P450 substrate binding preferences. Extrapolating from the models presented here, we postulate that the substantial protrusion of three residues into the CYP6Z2 catalytic site of the PEST and G3 strains significantly limits its substrate range. Whether mutations in these critical positions modulate the permethrin resistance reported in other CYP6Z2-overexpressing strains [e.g., Odumasy (15)] is not yet clear.
With CYP6Z1 from the susceptible G3 strain shown to be capable of metabolizing DDT and known amino acid variations in CYP6Z1 of the resistant RSP-ST strain not predicted to affect the DDT binding mode, the DDT resistance levels reported for the RSP-ST strain may possibly be due to the enhanced expression or stability of the CYP6Z1 protein in the RSP-ST strain or to an enhanced electron transfer rate (if the T346N variation affects interactions with P450 reductase). Although CYP6Z1 transcript levels have not yet been directly compared in the RSP-ST and G3 strains and resistance mechanisms may differ among strains, it is already known that CYP6Z1 transcript levels are higher in the resistant ZAN/U strain than in the susceptible Kisumu strain (11) as would be consistent with the first of these possibilities.
With the availability of these models and our P450 expression capabilities, clonings of the CYP6Z alleles expressed in other strains of An. gambiae provide us with the opportunity to differentiate critical catalytic site variants from superficial variants not affecting substrate specificity. For this, systematic analysis of allelic variants expressed in individual strains and other P450s reported to be overexpressed in resistant strains (CYP4C27, CYP4H15, and CYP12F1) will be essential for understanding the evolution of P450-mediated resistance mechanisms and designing strategies to moderate enhanced resistance. This study has made it clear that P450 metabolic profiles cannot be predicted simply from the primary sequences of related P450s. Three-dimensional representations of these proteins present us and other researchers with the opportunity to identify and test CYP6Z1-specific inhibitors capable of reducing DDT resistance levels in natural populations of this pest.
Materials and Methods
Sequence Alignments.
The sequences of CYP6Z1 (GenBank accession no. AF487535) from the RSP-ST strain of mosquito (41, 42) and CYP6Z2 (GenBank accession no. XP_317252) from the PEST strain (ref. 10 and http://p450.sophia.inra.fr) were compared with one another, and their fixed alignment was compared with the alignment of eight class II (eukaryotic) P450 sequences (24). Finding that human CYP3A4 (Protein Data Bank ID code 1TQN; ref. 26) shared the highest sequence identity with CYP6Z1 and CYP6Z2, the CYP3A4 sequence was then aligned with the CYP6Z pair, and a final alignment (Fig. 1) was created for use in modeling.
Molecular Modeling.
Homology models for CYP6Z1 and CYP6Z2 were built separately by using the HOMOLOGY function of MOE (Chemical Computing Group) based on the final alignment mentioned above. The heme coordinates were copied from the CYP3A4 template structure and explicitly included in the homology modeling process to generate 10 intermediate models coarsely energy-minimized in MOE. The best intermediate models with the highest MOE residue packing scores (−2.7706 for CYP6Z1 and −2.7530 for CYP6Z2) were then selected for further energy minimizations using the CHARMM22 force field (47) as distributed in MOE in the presence of the heme coordinates duplicated from the template structure.
The final substrate-free protein models were examined for their distributions of backbone torsion angles by using Ramachandran plots generated within MOE. The models were then tested by using Prosa II (version 3.0) analysis (Center for Applied Molecular Engineering, University of Salzburg, Salzburg, Austria) and Profiles3D analysis (Insight II Homology module; Accelrys). Normalized Z-score calculations, which determine the appropriateness of folding relative to other known protein structures at values >0.7, indicated that the CYP6Z1 and CYP6Z2 models have values of 0.81 and 0.82, respectively. Profiles3D scores calculated for the CYP6Z1 and CYP6Z2 models as a function of residue number showed positive compatibility scores throughout the models indicating that every residue is in a compatible structural environment. The overall qualities of the two models are demonstrated by a summed compatibility score for the CYP6Z1 model of 176.55, which lies between the lower bound (bad structures) of 97.83 and the upper bound (ideal structures) of 217.40, and for the CYP6Z2 model of 198.40, which lies between the lower bound of 97.42 and the upper bound of 216.48.
DDT, carbaryl, and xanthotoxin were docked within predicted catalytic sites by using the DOCK function of MOE. In preparation for docking, a singlet oxygen was attached to the heme plane by using the parameter assignments specified by Rupasinghe et al. (30). The initial positions of these compounds were set above the heme plane, and docking simulations were carried out by using the MMFF94s force field (31–36) and the simulated annealing conformation search method within the DOCK function. One hundred to 300 conformations for DDT and 100 conformations each for carbaryl and xanthotoxin were generated while keeping protein side chains fixed, and these were sorted in ascending order according to their total energy. Binding modes with the lowest total energies and the trichloromethyl group of DDT, carbons 4–7 on carbaryl, or the furan ring on xanthotoxin closest to the heme were chosen for second-round energy minimizations during which all protein side chains were allowed to move freely while the heme coordinates were fixed to avoid distortion of the heme plane.
Heterologous Expression and Metabolism.
Details on the cloning of CYP6Z1 and CYP6Z2, their coexpression with P450 reductase in Sf9 cells, and metabolism assays for DDT, carbaryl, and xanthotoxin are included in SI Methods.
Supplementary Material
Acknowledgments.
We thank Dr. May Berenbaum and Dr. Jerome Baudry for scientific comments on the manuscript. This research was funded by National Institutes of Health Grant R01 GM071826.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709249105/DCSupplemental.
References
- 1.Njoroge GN, Bussmann RW. Diversity and utilization of antimalarial ethnophytotherapeutic remedies among the Kikuyus (Central Kenya) J Ethnobiol Ethnomed. 2006;2:8–14. doi: 10.1186/1746-4269-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hemingway J, Bates I. Malaria: Past problems and future prospects. EMBO Rep. 2003;4:S29–S31. doi: 10.1038/sj.embor.embor841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osta MA, Christophides GK, Kafatos FC. Effects of mosquito genes on Plasmodium development. Science. 2004;303:2030–2032. doi: 10.1126/science.1091789. [DOI] [PubMed] [Google Scholar]
- 4.Hemingway J, Ranson H. Insecticide resistance in insect vectors of human disease. Annu Rev Entomol. 2000;45:371–391. doi: 10.1146/annurev.ento.45.1.371. [DOI] [PubMed] [Google Scholar]
- 5.Hemingway J, Field L, Vontas J. An overview of insecticide resistance. Science. 2002;298:96–97. doi: 10.1126/science.1078052. [DOI] [PubMed] [Google Scholar]
- 6.Hemingway J, Hawkes NJ, McCarroll L, Ranson H. The molecular basis of insecticide resistance in mosquitoes. Insect Biochem Mol Biol. 2004;34:653–665. doi: 10.1016/j.ibmb.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 7.Craig A, Kyes S, Ranson H, Hemingway J. Malaria parasite and vector genomes: Partners in crime. Trends Parasitol. 2003;19:356–362. doi: 10.1016/s1471-4922(03)00146-6. [DOI] [PubMed] [Google Scholar]
- 8.Hemingway J. Taking aim at mosquitoes. Nature. 2004;430:936. doi: 10.1038/430936a. [DOI] [PubMed] [Google Scholar]
- 9.Coetzee M, Horne DWK, Brooke BD, Hunt RH. DDT, dieldrin and pyrethroid insecticide resistance in African malaria vector mosquitoes: An historical review and implications for future malaria control in southern Africa. S Afr J Sci. 1999;95:215–218. [Google Scholar]
- 10.Holt RA, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- 11.David JP, et al. The Anopheles gambiae detoxification chip: A highly specific microarray to study metabolic-based insecticide resistance in malaria vectors. Proc Natl Acad Sci USA. 2005;102:4080–4084. doi: 10.1073/pnas.0409348102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott JG. Cytochromes P450 and insect resistance. Insect Biochem Mol Biol. 1999;29:757–777. doi: 10.1016/s0965-1748(99)00038-7. [DOI] [PubMed] [Google Scholar]
- 13.Nikou D, Ranson H, Hemingway J. An adult-specific CYP6 P450 gene is overexpressed in a pyrethroid-resistant strain of the malaria vector, Anopheles gambiae. Gene. 2003;318:91–102. doi: 10.1016/s0378-1119(03)00763-7. [DOI] [PubMed] [Google Scholar]
- 14.Strode C, Steen K, Ortelli F, Ranson H. Differential expression of the detoxification genes in the different life stages of the malaria vector Anopheles gambiae. Insect Mol Biol. 2006;15:523–530. doi: 10.1111/j.1365-2583.2006.00667.x. [DOI] [PubMed] [Google Scholar]
- 15.Muller P, Donnelly MJ, Ranson H. Transcription profiling of a recently colonised pyrethroid resistant Anopheles gambiae strain from Ghana. BMC Genomics. 2007;8:36. doi: 10.1186/1471-2164-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersen JF, Utermohlen JG, Feyereisen R. Expression of house fly CYP6A1 and NADPH-cytochrome P450 reductase in Escherichia coli and reconstitution of an insecticide-metabolizing P450 system. Biochemistry. 1994;33:2171–2177. doi: 10.1021/bi00174a025. [DOI] [PubMed] [Google Scholar]
- 17.Zhang M, Scott JG. Cytochrome b5 is essential for cytochrome P450 6D1-mediated cypermethrin resistance in LPR house flies. Pestic Biochem Physiol. 1996;55:150–156. doi: 10.1006/pest.1996.0044. [DOI] [PubMed] [Google Scholar]
- 18.Dunkov BC, et al. The Drosophila cytochrome P450 gene Cyp6a2: Structure, localization, heterologous expression, and induction by Phenobarbital. DNA Cell Biol. 1997;16:1345–1356. doi: 10.1089/dna.1997.16.1345. [DOI] [PubMed] [Google Scholar]
- 19.Sabourault C, et al. Overproduction of a P450 that metabolizes diazinon is linked to a loss-of-function in the chromosome 2 ali-esterase (MdαE7) gene in resistant houseflies. Insect Mol Biol. 2001;10:609–618. doi: 10.1046/j.0962-1075.2001.00303.x. [DOI] [PubMed] [Google Scholar]
- 20.Amichot M, et al. Point mutations associated with insecticide resistance in the Drosophila cytochrome P450 Cyp6a2 enable DDT metabolism. Eur J Biochem. 2004;271:1250–1257. doi: 10.1111/j.1432-1033.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Baudry J, Berenbaum MR, Schuler MA. Structural and functional evolution of insect CYP6B proteins: From specialist to generalist P450. Proc Natl Acad Sci USA. 2004;101:2939–2944. doi: 10.1073/pnas.0308691101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rupasinghe S, Wen Z, Chiu T-L, Schuler MA. Helicoverpa zea CYP6B8 and CYP321A1: Different molecular solutions to the problem of metabolizing plant toxins and insecticides. Protein Eng Des Sel. 2007;20:615–624. doi: 10.1093/protein/gzm063. [DOI] [PubMed] [Google Scholar]
- 23.Dai R, Pincus MR, Friedman FK. Molecular modeling of mammalian cytochrome P450s. Cell Mol Life Sci. 2000;57:487–499. doi: 10.1007/PL00000709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baudry J, Rupasinghe S, Schuler MA. Class-dependent sequence alignment strategy improves the structural and functional modeling of P450s. Protein Eng Des Sel. 2006;19:345–353. doi: 10.1093/protein/gzl012. [DOI] [PubMed] [Google Scholar]
- 25.Rupasinghe S, Duan H, Schuler MA. Molecular definitions of fatty acid hydroxylases in Arabidopsis thaliana. Proteins. 2007;68:279–293. doi: 10.1002/prot.21335. [DOI] [PubMed] [Google Scholar]
- 26.Yano JK, et al. The structure of human microsomal cytochrome P450 3A4 determined by x-ray crystallography to 2.05-Å resolution. J Biol Chem. 2004;279:38091–38094. doi: 10.1074/jbc.C400293200. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Berenbaum MR, Schuler MA. Molecular cloning and expression of CYP6B8: A xanthotoxin-inducible cytochrome P450 cDNA from Helicoverpa zea. Insect Biochem Mol Biol. 2000;30:75–84. doi: 10.1016/s0965-1748(99)00102-2. [DOI] [PubMed] [Google Scholar]
- 28.Edelsbrunner H, Facello M, Fu P, Liang J. Measuring proteins and voids in proteins. Proceedings of the Twenty-Eighth Annual Hawaii International Conference on System Sciences; Los Alamitos, CA: IEEE Comput Soc Press; 1995. pp. 256–264. [Google Scholar]
- 29.Wade RC, Winn PJ, Schlichting I, Sudarko A survey of active sites access channels in cytochromes P450. J Inorg Biochem. 2004;98:1175–1182. doi: 10.1016/j.jinorgbio.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 30.Rupasinghe S, Baudry J, Schuler MA. Common active site architecture and binding strategy of four phenylpropanoid P450s from Arabidopsis thaliana as revealed by molecular modeling. Protein Eng. 2003;16:721–731. doi: 10.1093/protein/gzg094. [DOI] [PubMed] [Google Scholar]
- 31.Halgren TA. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comp Chem. 1996;17:490–519. [Google Scholar]
- 32.Halgren TA. Merck molecular force field. II. MMFF94 van der Waals and electrostatic parameters for intermolecular interactions. J Comp Chem. 1996;17:520–552. [Google Scholar]
- 33.Halgren TA. Merck molecular force field. III. Molecular geometries and vibrational frequencies for MMFF94. J Comp Chem. 1996;17:553–586. [Google Scholar]
- 34.Halgren TA, Nachbar RB. Merck molecular force field. IV. Conformational energies and geometries for MMFF94. J Comp Chem. 1996;17:587–615. [Google Scholar]
- 35.Halgren TA, Nachbar RB. Merck molecular force field. V. Extension of MMFF94 using experimental data, additional computational data, and empirical rules. J Comp Chem. 1996;17:616–641. [Google Scholar]
- 36.Halgren TA. MMFF. VI. MMFF94s option for energy minimization studies. J Comp Chem. 1999;20:720–729. doi: 10.1002/(SICI)1096-987X(199905)20:7<720::AID-JCC7>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 37.Kitamura S, et al. Reductive metabolism of p,p′-DDT and o,p′-DDT by rat liver cytochrome P450. Drug Metab Dispos. 2002;30:113–118. doi: 10.1124/dmd.30.2.113. [DOI] [PubMed] [Google Scholar]
- 38.Tang J, Cao Y, Rose RL, Hodgson E. In vitro metabolism of carbaryl by human cytochrome P450 and its inhibition by chlorpyrifos. Chem Biol Inter. 2002;141:229–241. doi: 10.1016/s0009-2797(02)00074-1. [DOI] [PubMed] [Google Scholar]
- 39.Ivie GW, Bull DL, Beier RC, Pryor NW, Oertli EH. Metabolic detoxification: Mechanism of insect resistance to plant psoralens. Science. 1983;221:374–376. doi: 10.1126/science.221.4608.374. [DOI] [PubMed] [Google Scholar]
- 40.Binkowski TA, Naghibzadeh S, Liang J. CASTp: Computed Atlas of Surface Topography of proteins. Nucleic Acids Res. 2003;31:3352–3355. doi: 10.1093/nar/gkg512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vulule JM, et al. Elevated oxidase and esterase levels associated with permethrin tolerance in Anopheles gambiae from Kenyan villages using permethrin-impregnated nets. Med Vet Entomol. 1999;13:239–244. doi: 10.1046/j.1365-2915.1999.00177.x. [DOI] [PubMed] [Google Scholar]
- 42.Ranson H, et al. Molecular analysis of multiple cytochrome P450 genes from the malaria vector, Anopheles gambiae. Insect Mol Biol. 2002;11:409–418. doi: 10.1046/j.1365-2583.2002.00350.x. [DOI] [PubMed] [Google Scholar]
- 43.Danielson PB. The cytochrome P450 superfamily: Biochemistry, evolution and drug metabolism in humans. Curr Drug Metab. 2002;3:561–597. doi: 10.2174/1389200023337054. [DOI] [PubMed] [Google Scholar]
- 44.Haudenschild C, Schalk M, Karp F, Croteau R. Functional expression of regiospecific cytochrome P450 limonene hydroxylases from mint (Mentha spp.) in Escherichia coli and Saccharomyces cerevisiae. Arch Biochem Biophys. 2000;379:127–136. doi: 10.1006/abbi.2000.1864. [DOI] [PubMed] [Google Scholar]
- 45.Schalk M, Croteau R. A single amino acid substitution (F363I) converts the regiochemistry of the spearmint (−)-limonene hydroxylase from a C6- to a C3-hydroxylase. Proc Natl Acad Sci USA. 2000;97:11948–11953. doi: 10.1073/pnas.97.22.11948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wen Z, Rupasinghe S, Niu G, Berenbaum MR, Schuler MA. CYP6B1 and CYP6B3 of the black swallowtail (Papilio polyxenes): Adaptive evolution through subfunctionalization. Mol Biol Evol. 2006;23:2434–2443. doi: 10.1093/molbev/msl118. [DOI] [PubMed] [Google Scholar]
- 47.MacKerell AD, Jr, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.