Abstract
Cytosine methylation at CpG dinucleotides produces m5CpG, an epigenetic modification that is important for transcriptional regulation and genomic stability in vertebrate cells. However, m5C deamination yields mutagenic G·T mispairs, which are implicated in genetic disease, cancer, and aging. Human thymine DNA glycosylase (hTDG) removes T from G·T mispairs, producing an abasic (or AP) site, and follow-on base excision repair proteins restore the G·C pair. hTDG is inactive against normal A·T pairs, and is most effective for G·T mispairs and other damage located in a CpG context. The molecular basis of these important catalytic properties has remained unknown. Here, we report a crystal structure of hTDG (catalytic domain, hTDGcat) in complex with abasic DNA, at 2.8 Å resolution. Surprisingly, the enzyme crystallized in a 2:1 complex with DNA, one subunit bound at the abasic site, as anticipated, and the other at an undamaged (nonspecific) site. Isothermal titration calorimetry and electrophoretic mobility-shift experiments indicate that hTDG and hTDGcat can bind abasic DNA with 1:1 or 2:1 stoichiometry. Kinetics experiments show that the 1:1 complex is sufficient for full catalytic (base excision) activity, suggesting that the 2:1 complex, if adopted in vivo, might be important for some other activity of hTDG, perhaps binding interactions with other proteins. Our structure reveals interactions that promote the stringent specificity for guanine versus adenine as the pairing partner of the target base and interactions that likely confer CpG sequence specificity. We find striking differences between hTDG and its prokaryotic ortholog (MUG), despite the relatively high (32%) sequence identity.
Keywords: CpG site, DNA repair, G·T mismatch, deamination, 5-methylcytosine
Human thymine DNA glycosylase (hTDG) belongs to the uracil DNA glycosylase (UDG) superfamily of enzymes that share a common α/β fold and promote genomic integrity by removing mutagenic uracil bases from DNA (1, 2). Initiating the base excision repair pathway, these enzymes use a remarkable nucleotide-flipping mechanism to extrude damaged nucleobases from the DNA helix and cleave the base-sugar (N-glycosidic) bond, producing an abasic (or AP) site in the DNA (3). Together, hTDG and the Escherichia coli mismatch-specific uracil DNA glycosylase (eMUG) are the most thoroughly characterized members of the TDG/MUG family (4–6). These enzymes excise a variety of damaged bases (X), and typically exhibit a strong preference for lesions in G·X versus A·X pairs (7–12). Like its eukaryotic orthologs, hTDG (410 residues) contains a conserved catalytic core (residues 121–300) flanked by more divergent N- and C-terminal domains (6); the former enhances DNA binding and G·T repair activity to some extent (13, 14), and the latter contains a site for SUMO conjugation (K330), a modification that decreases the DNA-binding affinity of hTDG (15, 16).
A recent structure of the hTDG catalytic domain (residues 117–332, conjugated to SUMO-1) reveals strong similarity to the structure of eMUG (16, 17), consistent with the 32% amino acid sequence identity. Nevertheless, the specificity of these enzymes differs remarkably. They both act on G·U mispairs, but only hTDG has significant activity for G·T mispairs (5, 9). Specificity for damage at CpG dinucleotides also distinguishes hTDG from eMUG and the vast majority of other DNA glycosylases (7, 8, 12, 17). The sole exception, MBD4, recognizes G·T mispairs and other lesions with specificity for CpG sites (18–21). The CpG specificity suggests that the predominant biological substrate for hTDG may be G·T mismatches arising from m5C deamination (22), because cytosine methylation occurs selectively at CpG sites in vertebrate cells. Because hTDG excises thymine, it must also employ a stringent mechanism to avoid acting on the huge (millionfold) excess of undamaged A·T base pairs. We recently showed that hTDG activity is 18,000-fold lower for A·T versus G·T pairs and sharply reduced for other lesions in an A·X versus G·X context (12). However, the preferential excision of bases paired with G also demands a mechanism to minimize the excision of C from normal G·C pairs. Our recent studies indicate that specificity for G·T over G·C pairs is largely attributable to the greater base–sugar (N-glycosidic) bond stability for deoxycytidine versus deoxythymidine, rather than selective base recognition or an inability of hTDG to flip cytosine into its active site (11). Our findings here provide a structural basis of the specificity for G·T versus A·T pairs (and G·X versus A·X pairs) and for lesions that arise in a CpG sequence context.
Results and Discussion
Overall Structure.
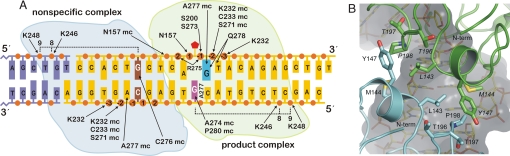
We obtained crystals of hTDG catalytic domain (hTDGcat, residues 111–308) bound to 22-bp DNA containing a tetrahydrofuran nucleotide (THF), a chemically stable mimic of the natural AP product. Previous studies showed that hTDG binds tightly to DNA containing G·THF, as it does to G·AP DNA (23). We solved the structure by molecular replacement using the structure of hTDG conjugated to SUMO-1 (16), and refined it to a crystallographic R factor of 23.0% and an Rfree of 27.6%, at a resolution of 2.8 Å [supporting information (SI) Table S1]. We found that hTDGcat crystallized in a 2:1 complex with the DNA, one subunit positioned at the abasic site (product complex), as expected, and the other at an undamaged or nonspecific (NS) site (Fig. 1 and Fig. S1). This was surprising, because 2:1 binding had not previously been reported for hTDG, eMUG, or UDG. Fig. 2 A provides a schematic overview of the enzyme–DNA interactions and the binding site for each hTDGcat subunit. In the 2:1 complex, the protein subunits form a symmetrical dimer interface, burying ≈290 Å2 of accessible surface area (ASA) per monomer (≈3% of total ASA) with no apparent hydrogen bonds or salt bridges (Fig. 2B). The small size of this interface indicates that the hTDGcat dimer is weak in the absence of DNA (24). This is confirmed by sedimentation velocity analytical ultracentrifugation experiments (Fig. S2), which show that hTDGcat is fully monomeric at a concentration of 118 μM. Thus, the initial DNA-binding event likely involves a monomer of hTDGcat. Residues contributing to the dimer interface, determined by buried surface area, include L143, M144, and Y147 of helix α1, and T196, T197, and P198 of the β2–α4 loop (Fig. 1). These residues are strictly conserved for vertebrate TDGs but not with TDG from Drosophila or fission yeast (Saccharomyces pombe) (10) or with eMUG (Fig. S3). Our structure raises the questions of whether 2:1 binding is observed in solution and whether it is required for catalytic activity.
Fig. 1.
Overview of the structure. hTDGcat binds the DNA in a 2:1 complex: one subunit at the abasic site (product complex) and the other at an undamaged site (nonspecific complex). DNA shown includes a full 22-bp duplex and part of the adjacent duplex joined by 3′ A/T overhangs (blue arrow, see Fig. 2A). The secondary structure is also shown with the amino acid sequence in Fig. S3. Overall, the two subunits are highly similar (rms deviation of 0.8 Å for Cα positions).
Fig. 2.
Schematic overview of the enzyme–DNA interactions and the dimer interface. (A) The 22-bp DNA is yellow with phosphates shown as orange circles. The adjoining DNA fragment (purple) shows contacts with K246 and K248 from the NS subunit. The arrows represent hydrogen bonds involving side-chain or main-chain (mc) atoms of the enzyme. In the product complex, the flipped abasic nucleotide (THF) is a red pentagon, the “opposing G” is magenta, and the “3′-G” is cyan. A277 intercalates the complementary strand, disrupting base-stacking interactions between the opposing G and its 5′ neighbor. Contacts involving N157, S273, and A274 for hTDGcat are topologically conserved with contacts in the eMUG product complex (17), and the N157, K232, S271, and S273 contacts are conserved with those in the UDG product complex (29). (B) Close-up view of the dimer interface, with the G·THF-bound subunit in green and the nonspecific subunit in cyan. The N termini of each subunit (T123) are indicated.
Biochemical Studies.
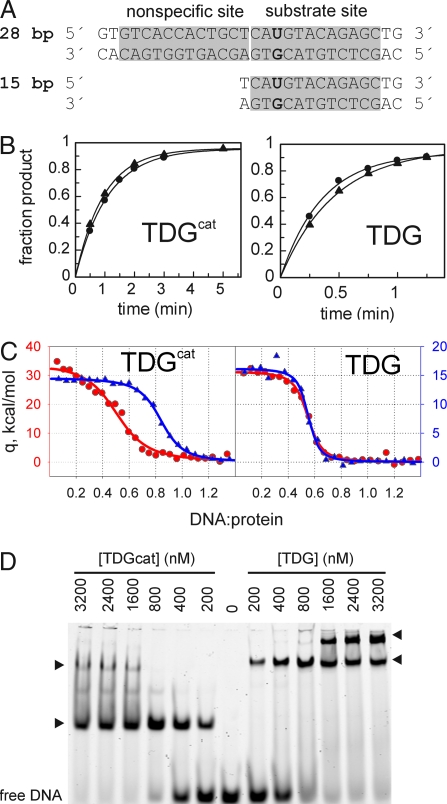
We performed single-turnover kinetics using two G·U-containing substrates (Fig. 3A), a 28-bp duplex (G·U28) that can accommodate all of the observed protein–DNA interactions for each hTDGcat subunit (see Fig. 2A) and a 15-bp substrate (G·U15) that lacks the entire binding site for the NS subunit. The activity of hTDGcat is identical for the G·U28 and G·U15 substrates, kmax = 1.0 ± 0.2 min−1 (Fig. 3B). Likewise, full-length hTDG exhibits nearly the same activity for G·U28, kmax = 2.9 ± 0.3 min−1, as for G·U15, kmax = 2.0 ± 0.3 min−1. Thus, the 2:1 complex observed in the crystal structure is not required for full catalytic activity. However, this does not exclude the possibility of 2:1 binding at substrate sites. The NS subunit might bind even if it is not needed for catalysis. Alternatively, the 2:1 complex might arise at abasic sites; hTDG binds tightly to its abasic DNA product (25).
Fig. 3.
Enzyme kinetics and binding studies. (A) G·U-containing DNA substrates used for single-turnover kinetics. The 28-bp (G·U28) DNA contains the nonspecific binding site as seen in the crystal structure, but the 15-bp (G·U15) does not. The THF28 and THF15 used for ITC and EMSA are identical to G·U28 and G·U15, except that THF replaces dU. (B) Progress curves and fitted data from representative single-turnover kinetics for hTDGcat or hTDG (2.5 μM) acting on G·U28 (●) or G·U15 (▴) (250 nM). (C) Isothermal titration calorimetry data for hTDG or hTDGcat binding to THF28 (red, ●) or THF-15 (blue, ▴). Note carefully that the scale for THF28 data (red, left axis) is twofold greater than the scale for THF15 data (blue, right axis) to illustrate the near doubling of ΔH for binding THF28 relative to THF15. (D) EMSA was performed with 200 nM THF28 (6-FAM labeled) and the enzyme concentrations shown. Arrowheads indicate the apparent 1:1 (Lower) and 2:1 (Upper) protein:DNA complexes. The 2:1 complex for hTDGcat (23 kDa for monomer) has mobility similar to that of the 1:1 complex for hTDG (46-kDa monomer).
We investigated the latter possibility using isothermal titration calorimetry (ITC), with DNA identical to G·U28 and G·U15 except that THF replaces dU. The ITC results (Fig. 3C and Table 1) indicate that binding of hTDG (and hTDGcat) to THF15 or THF28 is endothermic, thus driven by favorable entropy (TΔS > 0) that compensates for the large unfavorable enthalpy (ΔH > 0) (at 5°C), consistent with previous findings for proteins that distort DNA (26). It is important to note that in Fig. 3C, the scale for the THF28 data (red, left axis) is twofold greater than that for THF15 (blue, right axis), to illustrate the near doubling of ΔH for THF28 versus THF15 binding to either hTDG or hTDGcat. A similar (nearly twofold) change in TΔS is seen for THF28 relative to THF15. Our results indicate that hTDG and hTDGcat form a 2:1 complex with THF28, as seen in the crystal structure, whereas only one enzyme subunit interacts with THF15. Although the n = 0.5 (DNA/protein) stoichiometry observed for hTDG binding to THF15 suggests a protein dimer, the thermodynamic and structural results indicate one hTDG subunit interacting with DNA. Previous ITC results for a DNA glycosylase (Fpg) that is known to bind DNA with 1:1 stoichiometry, ΔH = 14.2 kcal/mol and TΔS = 23.5 kcal/mol (5°C) (27), are remarkably similar to our results for THF15, consistent with our conclusion that THF15 interacts with one enzyme subunit, and THF28 interacts with two. An electrophoretic mobility shift assay (EMSA) (Fig. 3D) indicates that hTDG and hTDGcat can form 1:1 or 2:1 complexes with THF28 DNA. The 1:1 complex is more stable than the 2:1 (at least under EMSA conditions), particularly for hTDGcat. Together, the ITC and EMSA results suggest that the protein dimer may be stronger for hTDG than for hTDGcat. Intriguingly, the close proximity of the N termini of hTDGcat seen in the crystal structure (Fig. 2B) suggests that this might be due to interactions between the N-terminal domains of full-length hTDG. Absent a clear role in catalysis, it is conceivable that the 2:1 complex, if adopted in vivo, could be important for some other function of hTDG. The enzyme is known to interact with proteins involved in transcriptional regulation, with the Dnmt3a/b methyltransferases and the Rad9-Rad1-Hus1 cell cycle checkpoint complex (6, 14, 28). Additional studies are needed to fully explore conditions that favor 2:1 binding and its potential biological role(s).
Table 1.
Thermodynamic parameters for hTDG and hTDGcat binding to abasic DNA
| Enzyme | DNA | ΔH, kcal/mol | TΔS, kcal/mol | ΔG, kcal/mol | N |
|---|---|---|---|---|---|
| hTDGcat | THF28 | 34 ± 1 | 43 ± 1 | −8.9 ± 0.1 | 0.58 ± 0.04 |
| hTDGcat | THF15 | 14.6 ± 0.1 | 24.2 ± 0.2 | −9.6 ± 0.1 | 0.81 ± 0.05 |
| hTDG | THF28 | 29.5 ± 0.9 | 39.9 ± 0.9 | −10.23 ± 0.03 | 0.52 ± 0.03 |
| hTDG | THF15 | 17.3 ± 0.4 | 24.8 ± 0.7 | −10.34 ± 0.06 | 0.50 ± 0.02 |
Values represent mean ± SD determined from at least three independent experiments.
Overall Enzyme–DNA Interactions in the Product Complex.
As shown in Fig. 4 and Fig. S1, hTDGcat dramatically distorts the DNA in the product complex, predominantly at the abasic (THF) site. The abasic nucleotide is flipped completely out of the helix and into the active site, and the DNA helix is bent by ≈43°. Notably, human UDG imposes a similar 45° kink in the DNA (3, 29), but eMUG imparts little DNA bending (17, 30). hTDGcat forms numerous contacts with the abasic strand, involving phosphodiester groups flanking the abasic nucleotide, the abasic sugar, and both guanines of the CpG site, burying a surface area of 1,107 Å2 and 1,056 Å2 for the DNA and protein, respectively. Although the overall structure of hTDGcat bound to G·THF DNA closely resembles that of eMUG bound to G·AP DNA (17), with an rms deviation of 1.4 Å for Cα positions, large and unexpected differences in the protein-DNA interactions are observed, as detailed below.
Fig. 4.
Close-up view of the hTDGcat-G·THF product complex. The enzyme is represented as a semitransparent surface, showing key residues in stick format. DNA is represented in stick format with the abasic strand colored green and the complementary strand colored yellow (oxygen red, nitrogen blue, phosphorus orange). Phosphates are labeled according to their position with respect to the target site (THF). Hydrogen bond interactions between the R275 side chain and the P2 and P-1 phosphates are indicated by black dashed lines. The orange dashed line indicates the “phosphate pinch,” where the distance between P2 and P-1 is reduced to 7.8 Å from an average of 12.7 Å for the DNA overall.
As shown in Fig. 2A, hTDGcat contacts two backbone phosphates in the complementary strand at positions 8 and 9 on the 5′ side of the G·THF site, by using the side chains of K246 and K248. This explains the previous observation from biochemical (DNase I footprinting) studies that hTDG “protects” 10 nucleotides positioned 5′ to the G of a G·U site (31). Moreover, the contacts are consistent with our observation that maximal catalytic activity is reduced by ≈10-fold for DNA containing 6 bp as compared with 9 bp on the 5′ side of the G in G·U substrate DNA (data not shown). Notably, K246 and K248 are strictly conserved in vertebrate TDGs, but not eMUG or UDG. Accordingly, these long-range phosphate contacts are not seen for DNA complexes of eMUG or UDG (17, 32) and are indeed rare for any DNA glycosylase (3).
As shown in Figs. 2A and 4, hTDGcat contacts five backbone phosphates of the target strand in the product complex, two located 5′ of the target (THF) site (P1 and P2) and three on its 3′ side (P-1, P-2, and P-3). The hTDG residues that provide these interactions and the phosphate nomenclature are given in Fig. 2A. As shown in Fig. 4, the phosphate belonging to the abasic nucleotide (P1) is displaced from the DNA helix and resides deep within the active-site pocket. Additionally, the phosphates flanking the abasic nucleotide (P2 and P-1) are compressed together by nearly 5 Å. This “phosphate pinch” was previously observed for UDG and eMUG and is thought to promote nucleotide flipping (17, 29, 30, 32). The phosphate pinch may also contribute to the chemical step by optimally positioning the phosphates relative to the AP sugar. Studies on UDG show that at least three flanking phosphates (P2, P1, and P-1, and perhaps P-2) provide electrostatic stabilization to the cationic sugar and repel the anionic leaving group in the transition state of the reaction, accounting for perhaps half of the observed enzymatic rate enhancement (3, 33, 34). Importantly, the distances between these phosphates and C1′ of the abasic nucleotide in our structure are nearly identical to those observed for UDG (32, 34). Moreover, hTDGcat fully buries the P-1 and P-2 phosphates and reduces the accessible surface area for P1 and P2 by 9- and 4-fold, respectively, enhancing their catalytic effect.
Strikingly, the R275 side chain deeply penetrates the DNA minor groove, plugging the space vacated by the flipped abasic nucleotide and contacting both the P2 and P-1 phosphates (Fig. 4). The corresponding “plug” for eMUG and hUDG is a Leu side chain, which is thought to “push” the target nucleotide into the active site (29, 32) or at least stabilize its extrahelical conformation (35). For hTDG, the electrostatic contacts from R275 to P2 and P-1 may enhance its promotion of nucleotide flipping and increase DNA binding affinity by stabilizing the insertion of the side chain within the DNA helix and by enforcing the phosphate pinch.
Importantly, previous sequence alignments of hTDG and eMUG suggested an entirely different role for R275; it was thought to intercalate the complementary strand adjacent to the guanine that pairs with the target base (opposing G in Fig. 4), as does R146 of eMUG (10, 17, 30). However, a new structure-based alignment (Fig. S3) reveals that A277, not R275, is topologically conserved with R146 of eMUG. As discussed below, A277 does invade the complementary strand at the opposing G. These two residues are part of a loop (β5-α6, Fig. 4) that inserts into the helix and serves many important roles in substrate recognition and catalysis. Strikingly, this “insertion loop” accounts for 67% of the buried surface area for hTDGcat in the product complex. The insertion loop is absolutely conserved within vertebrate TDGs, but poorly conserved with eMUG (Fig. S3). For eMUG, the “insertion loop” residues were identified from crystal structures as 139-PNPSGLSR-146 (17, 30), and previous alignments indicated that the corresponding hTDG residues were 268-VMPSSSAR-275 (2, 36). However, our structure establishes that the corresponding hTDG residues are actually 270-PSSSARCA-277 (Fig. S3), an important distinction with implications for some previous conclusions about the catalytic role of these residues. For example, M269 was implicated in substrate recognition based on its previous alignment with a key His residue in UDG (36). However, an M269H mutation produced only a modest decrease in substrate binding (36), and we find that M269 does not contact the DNA in the hTDGcat product complex nor does it appear likely to contact a nucleobase flipped into the active site in an enzyme-substrate complex. However, the residue that replaces M269 in the new alignment, S271, does contact the P-2 phosphate in the hTDGcat product complex, as indicated in Fig. 2A.
The flipped abasic nucleotide is stabilized by many interactions in the active-site pocket (Fig. 4). The edge of the THF ring is surrounded by I139, N140, and G142 from the highly conserved motif 138-GINPG-142 (17). Residues 198–200 cover the face of the abasic nucleotide and appear to pose a barrier to its retrograde flipping back into the helix. Removal of N140 by site-directed mutagenesis showed that it plays a key role (36), likely by positioning the nucleophilic water molecule for attack at the deoxyribose C1′ upon rupture of the glycosidic bond and/or by providing electrostatic stabilization to the positively charged deoxyribose in the transition state of the reaction (2, 3, 36). Accordingly, C1′ of THF is proximal (≈3.2 Å) to both the side chain and backbone carbonyl oxygens of N140 (Fig. 4). Although it has been suggested that the very slow catalytic turnover of hTDG (7, 8, 12) is attributable largely to nonspecific DNA interactions involving the N-terminal ≈120 residues (13), the catalytic core (hTDGcat) is also strongly product inhibited (6, 14, 37). The extensive interactions observed here with the abasic nucleotide and backbone phosphates of both DNA strands, including the R275 side-chain contacts, offer a structural explanation for the slow turnover of hTDGcat and hTDG.
Enzyme–DNA Interactions in the Nonspecific Complex.
As shown in Fig. 2A and Fig. S4, the NS subunit interacts predominantly with the complementary strand, and the contacts are less extensive than for the product complex. Although a nucleotide is not flipped into the active site, the P-2 and P-3 phosphate contacts seen in the product complex are present in the NS complex, as are the long-range contacts with K246 and K248. This allows identification of the “target” site in the NS complex (colored brown in Fig. 2A), which is displaced by 5 bp from the central G·THF site. Although the “target” happens to be cytosine for the DNA used here, there is no evidence that the NS complex is specific for G·C at this position. The phosphate contacts at P1, P2, and P-1 are absent in the NS complex, and, consistent with the absence of nucleotide flipping, the phosphates flanking the target nucleotide are not “pinched,” and the insertion loop does not penetrate the minor groove. Nevertheless, the NS complex buries a surface area of 705 Å2 and 706 Å2 for the DNA and protein, respectively (35% less than the product complex), and the DNA helix is bent by ≈25°. This significant deformation seems partially attributable to the insertion loop, which presses against the minor groove, with A277 disrupting the stacking between the G at the target site and its 5′ neighboring base.
Specificity for Guanine as the Pairing Partner of the Target Base.
Returning to the product complex, Fig. 5 shows how the insertion loop provides specificity for guanine versus adenine as the pairing partner of the target base, i.e., for G·X versus A·X base pairs. Notably, the opposing G remains in the helix, stacked with its 3′ neighbor. However, the insertion loop (predominantly A277) displaces the 5′ cytosine base and forms a cleft that surrounds the opposing G at its 5′ face and its Watson–Crick and minor-groove edges. The N1H and N2H2 of the opposing G are contacted by the backbone oxygens of A274 and P280; the A274 interactions are topologically conserved with eMUG, but the P280 contact is not. These interactions with the Watson–Crick regions of the opposing G can be formed only when the target nucleotide is flipped into the active site, and they are not compatible with adenine. In addition, these interactions may stabilize the insertion loop within the DNA helix, enhancing the lifetime of a substrate nucleotide flipped into the active site. Notably, the enzyme does not contact N7 of the opposing G, consistent with previous biochemical (methylation interference) studies (31).
Fig. 5.
Recognition of the opposing G and the CpG context. In the product complex, hTDGcat contacts the guanine positioned 3′ to the abasic nucleotide (3′-G, cyan) and the guanine across from the (THF) target site (i.e., the opposing G, magenta). hTDGcat does not contact the cytosine partner of the 3′-G. The enzyme and DNA are rendered and colored as in Fig. 4. Dashed lines represent hydrogen bonds indicated by the structure. The N2H2 and N1H of the opposing G are contacted by the backbone oxygen of A274 and P280, and the N2H2 of the 3′-G is contacted by the Q278 side chain carbonyl and the A277 backbone amide (N–H bond vector nearly perpendicular to plane of 3′-G base).
Specificity for Damage Located at CpG Sites.
As shown in Fig. 5, the insertion loop also contributes to the CpG-sequence specificity of hTDG, an unusual capability that is not evident in eMUG (17, 30). Previous work shows that hTDG activity is maximal for G·T and other G·X lesions having a 5′ C·G pair, i.e., CpG·X (7, 8, 12). The effect of altering the 5′-neighboring base pair is particularly large for G·T mispairs, where the maximal excision rate decreases by 37-, 96-, and 582-fold for TpG·T, GpG·T, and ApG·T, respectively, compared with CpG·T (8, 12). As shown in Fig. 5, hTDGcat contacts the guanine positioned 3′ to the target nucleotide ( 5′-CpG-3′/5′-XpG-3′). Strikingly, hTDGcat does not contact the cytosine positioned 5′ to the opposing G (5′-CpG-3′/5′-XpG-3′), indicating that hTDG cannot discern the methylation status of a CpG site, consistent with previous biochemical findings (31). The insertion loop (R275, C276, A277, Q278) provides a cleft that interacts with the 5′ face and minor-groove edge of the 3′-G. The 3′-G remains stacked with the 3′-flanking base and hydrogen bonded to its cytosine partner (the 5′-C). The major-groove edge of the 3′-G is proximal to the K201 side chain (Figs. 4 and 5), consistent with methylation interference studies indicating that N7 is protected by hTDG (31). The exocyclic N2H2 of the 3′-G is contacted by the A277 backbone and the Q278 side chain. The interactions formed with the 3′-G promote nucleotide flipping and/or the chemical step of the reaction (12), likely by stabilizing the insertion loop within the DNA helix and increasing the lifetime of a substrate nucleotide flipped into the active site. K201 and Q278 are absolutely conserved for vertebrate TDGs, which require CpG-sequence specificity to recognize G·T lesions caused by m5C deamination, but they are not conserved in TDG from fission yeast or in eMUG (Fig. S3), which have no such requirement.
Materials and Methods
Materials.
Full-length hTDG was purified as described (12). The 23-kDa catalytic domain of human TDG (hTDGcat, residues 111–308) was expressed and purified essentially as described (11, 12) (SI Text). DNA was synthesized, purified, and hybridized as described (12) (SI Text).
Crystal Growth, Data Collection, and Structure Determination.
Crystals were grown and prepared for x-ray diffraction as described (SI Text). X-ray diffraction data were collected at beamline 9-1 of the Stanford Synchrotron Radiation Laboratory and processed with DENZO and SCALEPACK (38) (Table S1). The structure of the hTDGcat–DNA complex was solved at 2.8 Å resolution by molecular replacement by using Phaser (39) and the structure of hTDG (117–332) conjugated to SUMO-1 as a search model (PDB ID code 1WYW, SUMO-1 subunit removed) (16). The DNA molecule was built manually into the difference density. The structure was subjected to simulated annealing and initial refinement with CNS (40), followed by restrained refinement with TLS in Refmac (41). Model building was done with Coot (42). Solvent-accessible surface area calculations were performed by using the program GETAREA (43). The structural figures were prepared with PyMOL (www.pymol.org).
Biochemical Studies.
Single-turnover kinetics were performed as described (11, 12). ITC experiments were performed by using a MicroCal VP-ITC instrument. Protein and DNA samples were dialyzed versus ITC buffer [10 mM Tris·HCl (pH 7.5), 0.1 M NaCl, 0.5 mM TCEP-HCl] and degassed. Experiments comprised an initial 2-μl injection followed by a series of 10-μl injections of DNA (15–30 μM) into a 1.4-ml volume of enzyme (2–4.5 μM) maintained at 5°C. Data were analyzed by using python scripts with improved baseline estimates in raw heat data and automatic evaluation of dilution heat as a fitting parameter. The stoichiometry (N), free energy (ΔG), and enthalpy of binding (ΔH) were determined as independent parameters, and the entropy of binding (ΔS) was determined separately by replacing ΔG with it as an independent parameter. For the EMSA, THF28 (with 6-FAM at 3′ end of the THF strand) was incubated with varying amounts of hTDG or hTDGcat (200–3,200 nM) for 30 min at room temperature. Samples (10 μl) were loaded onto a 10% native polyacrylamide gel that was cast with 0.5× TBE plus 5% glycerol and run with 0.5× TBE for 160 min, 100 V, at 4°C. Gels were imaged by using a Typhoon 9400 (GE Healthcare) using the blue-excited (488 nm) fluorescence mode.
Supplementary Material
Acknowledgments.
We thank Dr. E. Toth, Dr. A. Gnatt, Dr. X. Hu, and T. Charpentier for discussions and assistance with optimizing crystals; L. Eisele (NY State Department of Health) for collecting sedimentation equilibrium analytical ultracentrifugation data, and Dr. P. Schär (University of Basel) for providing an expression plasmid for hTDGcat. This work was supported by National Institutes of Health (NIH) Grant GM072711 (to A.C.D.), the University of Maryland Greenebaum Cancer Center, the University of Maryland Baltimore X-ray Crystallography Core, and the Biochemistry Core of the Wadsworth Center, NY State Department of Health. X-ray data were collected at the Stanford Synchrotron Radiation Laboratory (SSRL), a national user facility operated by Stanford University on behalf of the Department of Energy (DOE). The SSRL Structural Molecular Biology Program is supported by the DOE and the NIH.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (accession number 2RBA).
This article contains supporting information online at www.pnas.org/cgi/content/full/0711061105/DCSupplemental.
References
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Pearl LH. Structure and function in the uracil-DNA glycosylase superfamily. Mutat Res. 2000;460:165–181. doi: 10.1016/s0921-8777(00)00025-2. [DOI] [PubMed] [Google Scholar]
- 3.Stivers JT, Jiang YL. A mechanistic perspective on the chemistry of DNA repair glycosylases. Chem Rev. 2003;103:2729–2759. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- 4.Wiebauer K, Jiricny J. In vitro correction of G. T mispairs to G.C pairs in nuclear extracts from human cells. Nature. 1989;339:234–236. doi: 10.1038/339234a0. [DOI] [PubMed] [Google Scholar]
- 5.Gallinari P, Jiricny J. A new class of uracil-DNA glycosylases related to human thymine-DNA glycosylase. Nature. 1996;383:735–738. doi: 10.1038/383735a0. [DOI] [PubMed] [Google Scholar]
- 6.Cortazar D, Kunz C, Saito Y, Steinacher R, Schar P. The enigmatic thymine DNA glycosylase. DNA Repair (Amsterdam) 2007;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 7.Sibghat U, et al. Base analog and neighboring base effects on substrate specificity of recombinant human G:T mismatch-specific thymine DNA-glycosylase. Biochemistry. 1996;35:12926–12932. doi: 10.1021/bi961022u. [DOI] [PubMed] [Google Scholar]
- 8.Waters TR, Swann PF. Kinetics of the action of thymine DNA glycosylase. J Biol Chem. 1998;273:20007–20014. doi: 10.1074/jbc.273.32.20007. [DOI] [PubMed] [Google Scholar]
- 9.O'Neill RJ, et al., editors. Mismatch uracil glycosylase from Escherichia coli: A general mismatch or a specific DNA glycosylase? J Biol Chem. 2003;278:20526–20532. doi: 10.1074/jbc.M210860200. [DOI] [PubMed] [Google Scholar]
- 10.Hardeland U, Bentele M, Jiricny J, Schar P. The versatile thymine DNA-glycosylase: A comparative characterization of the human, Drosophila and fission yeast orthologs. Nucleic Acids Res. 2003;31:2261–2271. doi: 10.1093/nar/gkg344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bennett MT, et al. Specificity of human thymine DNA glycosylase depends on N-glycosidic bond stability. J Am Chem Soc. 2006;128:12510–12519. doi: 10.1021/ja0634829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan MT, Bennett MT, Drohat AC. Excision of 5-halogenated uracils by human thymine DNA glycosylase: Robust activity for DNA contexts other than CpG. J Biol Chem. 2007;282:27578–27586. doi: 10.1074/jbc.M704253200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steinacher R, Schar P. Functionality of human thymine DNA glycosylase requires SUMO-Regulated changes in protein conformation. Curr Biol. 2005;15:616–623. doi: 10.1016/j.cub.2005.02.054. [DOI] [PubMed] [Google Scholar]
- 14.Guan X, et al. The human checkpoint sensor Rad9-Rad1-Hus1 interacts with and stimulates DNA repair enzyme TDG glycosylase. Nucleic Acids Res. 2007;35:6207–6218. doi: 10.1093/nar/gkm678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardeland U, Steinacher R, Jiricny J, Schar P. Modification of the human thymine–DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 2002;21:1456–1464. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baba D, et al. Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature. 2005;435:979–982. doi: 10.1038/nature03634. [DOI] [PubMed] [Google Scholar]
- 17.Barrett TE, et al. Crystal structure of a G:T/U mismatch-specific DNA glycosylase: Mismatch recognition by complementary-strand interactions. Cell. 1998;92:117–129. doi: 10.1016/s0092-8674(00)80904-6. [DOI] [PubMed] [Google Scholar]
- 18.Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 19.Bellacosa A, et al. MED1, a novel human methyl-CpG-binding endonuclease, interacts with DNA mismatch repair protein MLH1. Proc Natl Acad Sci USA. 1999;96:3969–3974. doi: 10.1073/pnas.96.7.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Millar CB, et al. Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science. 2002;297:403–405. doi: 10.1126/science.1073354. [DOI] [PubMed] [Google Scholar]
- 21.Wu P, et al. Mismatch repair in methylated DNA. Structure and activity of the mismatch-specific thymine glycosylase domain of methyl-CpG-binding protein MBD4. J Biol Chem. 2003;278:5285–5291. doi: 10.1074/jbc.M210884200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abu M, Waters TR. The main role of human thymine-DNA glycosylase is removal of thymine produced by deamination of 5-methylcytosine and not removal of ethenocytosine. J Biol Chem. 2003;278:8739–8744. doi: 10.1074/jbc.M211084200. [DOI] [PubMed] [Google Scholar]
- 23.Scharer OD, Nash HM, Jiricny J, Laval J, Verdine GL. Specific binding of a designed pyrrolidine abasic site analog to multiple DNA glycosylases. J Biol Chem. 1998;273:8592–8597. doi: 10.1074/jbc.273.15.8592. [DOI] [PubMed] [Google Scholar]
- 24.Bahadur RP, Chakrabarti P, Rodier F, Janin J. A dissection of specific and nonspecific protein–protein interfaces. J Mol Biol. 2004;336:943–955. doi: 10.1016/j.jmb.2003.12.073. [DOI] [PubMed] [Google Scholar]
- 25.Waters TR, Gallinari P, Jiricny J, Swann PF. Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J Biol Chem. 1999;274:67–74. doi: 10.1074/jbc.274.1.67. [DOI] [PubMed] [Google Scholar]
- 26.Jen-Jacobson L, Engler LE, Jacobson LA. Structural and thermodynamic strategies for site-specific DNA binding proteins. Structure (London) 2000;8:1015–1023. doi: 10.1016/s0969-2126(00)00501-3. [DOI] [PubMed] [Google Scholar]
- 27.Minetti CA, et al. Energetics of lesion recognition by a DNA repair protein: Thermodynamic characterization of formamidopyrimidine–glycosylase (Fpg) interactions with damaged DNA duplexes. J Mol Biol. 2003;328:1047–1060. doi: 10.1016/s0022-2836(03)00365-6. [DOI] [PubMed] [Google Scholar]
- 28.Li YQ, Zhou PZ, Zheng XD, Walsh CP, Xu GL. Association of Dnmt3a and thymine DNA glycosylase links DNA methylation with base-excision repair. Nucleic Acids Res. 2007;35:390–400. doi: 10.1093/nar/gkl1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parikh SS, et al. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J. 1998;17:5214–5226. doi: 10.1093/emboj/17.17.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrett TE, et al. Crystal structure of a thwarted mismatch glycosylase DNA repair complex. EMBO J. 1999;18:6599–6609. doi: 10.1093/emboj/18.23.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scharer OD, Kawate T, Gallinari P, Jiricny J, Verdine GL. Investigation of the mechanisms of DNA binding of the human G/T glycosylase using designed inhibitors. Proc Natl Acad Sci USA. 1997;94:4878–4883. doi: 10.1073/pnas.94.10.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slupphaug G, et al. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- 33.Dinner AR, Blackburn GM, Karplus M. Uracil-DNA glycosylase acts by substrate autocatalysis. Nature. 2001;413:752–755. doi: 10.1038/35099587. [DOI] [PubMed] [Google Scholar]
- 34.Jiang YL, Ichikawa Y, Song F, Stivers JT. Powering DNA repair through substrate electrostatic interactions. Biochemistry. 2003;42:1922–1929. doi: 10.1021/bi027014x. [DOI] [PubMed] [Google Scholar]
- 35.Jiang YL, Kwon K, Stivers JT. Turning on uracil-DNA glycosylase using a pyrene nucleotide switch. J Biol Chem. 2001;276:42347–42354. doi: 10.1074/jbc.M106594200. [DOI] [PubMed] [Google Scholar]
- 36.Hardeland U, Bentele M, Jiricny J, Schar P. Separating substrate recognition from base hydrolysis in human thymine DNA glycosylase by mutational analysis. J Biol Chem. 2000;275:33449–33456. doi: 10.1074/jbc.M005095200. [DOI] [PubMed] [Google Scholar]
- 37.Tini M, et al. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol Cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- 38.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 39.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 40.Brunger AT, et al. Crystallography NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 41.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Fraczkiewicz R, Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J Comput Chem. 1998;19:319–333. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.