Abstract
Animal eyes are morphologically diverse. Their assembly, however, always relies on the same basic principle, i.e., photoreceptors located in the vicinity of dark shielding pigment. Cnidaria as the likely sister group to the Bilateria are the earliest branching phylum with a well developed visual system. Here, we show that camera-type eyes of the cubozoan jellyfish, Tripedalia cystophora, use genetic building blocks typical of vertebrate eyes, namely, a ciliary phototransduction cascade and melanogenic pathway. Our findings indicative of parallelism provide an insight into eye evolution. Combined, the available data favor the possibility that vertebrate and cubozoan eyes arose by independent recruitment of orthologous genes during evolution.
Keywords: evolution, gene, opsin, photoreceptor, cnidaria
The assembly of diverse animal eyes requires two fundamental building blocks, photoreceptors and dark shielding pigment. The function of photoreceptors is to convert light (stream of photons) into intracellular signaling. The photoreceptor cells (PRCs) are classified into two distinct types: rhabdomeric, characteristic of vision in invertebrate eyes; and ciliary, characteristic of vision in vertebrate eyes (1). In both ciliary and rhabdomeric PRCs, the seven-transmembrane receptor (opsin) associates with retinal to constitute a functional photosensitive pigment. Each photoreceptor type uses a separate phototransduction cascade. Rhabdomeric photoreceptors employ r-opsins and a phospholipase C cascade, whereas ciliary photoreceptors use c-opsins and a phosphodiesterase (PDE) cascade (2, 3). In general, the dark pigment reduces photon scatter and orients the direction optimally sensitive to light. The biochemical nature of the dark pigment appears more diverse than the phototransduction cascades used by the PRCs. Vertebrate eyes use melanin as their exclusive dark pigment. However, among invertebrates, pterins constitute the eye pigment in the polychaete Platynereis dumerilii (4), pterins and ommochromes are accumulated in eyes of Drosophila (5), and melanin is found rarely such as in the inverse cup-like eyes of the planarian, Dugesia (6).
Cnidaria, the likely sister group to the Bilateria, constitute the earliest branching phylum containing a well developed visual system. For example, Cubozoa (known as “box jellyfish”) have camera-type eyes with cornea, lens, and retina; unexpectedly, the cubozoan retina has ciliated PRCs that are typical for vertebrate eyes (7–9). Cubomedusae are active swimmers that are able to make directional changes in response to visual stimuli (10). The cubozoan jellyfish, Tripedalia cystophora (Fig. 1A), has four sensory structures called rhopalia that are equally spaced around the bell. In addition to two camera-type lens containing eyes at right angles to one another, each rhopalium has two pit-shaped and two slit-shaped pigment cup eyes (Fig. 1B). Thus, with six eyes located on each rhopalium, Tripedalia has 24 eyes altogether. Because the visual fields of individual eyes of the rhopalium partly overlap, Tripedalia (like other Cubomedusae) has an almost complete view of its surroundings. The lens-containing Tripedalia eyes have sophisticated visual optics as do advanced bilaterian phyla (11).
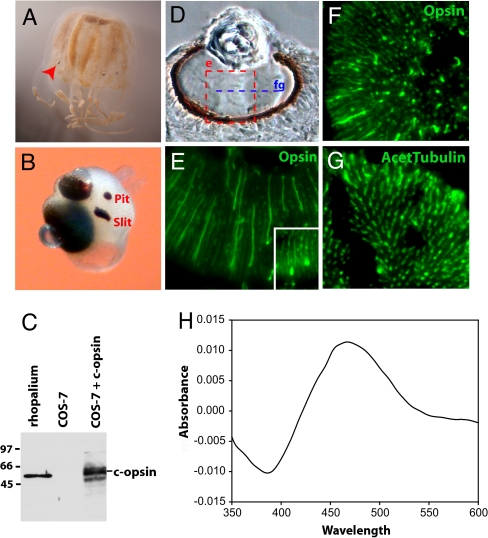
Fig. 1.
Ciliary opsin is the functional photopigment of Tripedalia camera-type eyes. (A) Tripedalia medusae with rhopalia (arrowhead). (B) Rhopalium with two lens-containing camera-type eyes, a slit and a pit eye. Another set of slit and pit eyes is symmetrically located on the other side of the rhopalium. (C) Specificity of an anti-Tripedalia c-opsin antibody tested by Western blotting with protein extracts from rhopalia and COS-7 cells transiently transfected with Tripedalia c-opsin cDNA. (D) Bright-field section through the large camera-type eye. The area shown in E is boxed. The plane of sectioning in F and G is indicated (fg). (E–G) Immunohistochemical staining (green) of Tripedalia retina using antibodies to c-opsin (E and F) or acetylated tubulin (G). (E Inset) Immunohistochemical staining for c-opsin in the small camera-type eye. (H) Dark–light difference absorption spectrum of the reconstituted Tripedalia c-opsin photopigment. The photopigment reconstituted with 11-cis-retinal forms a functional photopigment most sensitive to blue–green light.
In the present work, we characterize genes required for the assembly of camera-type eyes in Tripedalia. We show that the genetic building blocks typical of vertebrate eyes, namely ciliary opsin and the melanogenic pathway, are used by the cubozoan eyes. Although our findings of unsuspected parallelism are consistent with either an independent origin or common ancestry of cubozoan and vertebrate eyes, we believe the present data favor the former alternative.
Results
Ciliary Opsin Is Expressed in Camera-Type Eyes of Tripedalia.
We screened an expressed sequence tag (EST) library derived from rhopalia of Tripedalia to identify the jellyfish genes that are involved in vision; orthologues of other invertebrates and vertebrates were identified by phylogenetic analysis. Of the four opsin types present at the base of the bilaterians [rhabdomeric (r-opsins), ciliary (c-opsin), Go-opsins, and peropsin/RGR (12–14)], the Tripedalia opsin EST clustered with the c-opsins, an orthology consistent with the conservation of the characteristic stretch of deduced amino acids between the transmembrane domain VII and cytoplasmic tail [supporting information (SI) Fig. S1]. This region includes the c-opsin fingerprint tripeptide NR/KQ (NRS in Tripedalia) that is critical for coupling to the downstream phototransduction cascade through interaction with a GTP-binding protein subunit Gαt in the vertebrate rods and cones (15). An antibody generated against Tripedalia c-opsin recognized a single electrophoretic band in protein extracts prepared from rhopalia and COS-7 cells transfected with c-opsin cDNA (Fig. 1C). Camera-type eyes of adult jellyfish (Fig. 1D) were immunostained with an anti-c-opsin antibody. The c-opsin localized in the retinal ciliated PRCs of both complex eyes (Fig. 1 E and F) in a pattern resembling that by staining with anti-acetylated tubulin antibody (Fig. 1G), which specifically labels stabilized microtubules in axons and cilia (13).
Spectral Sensitivity of Tripedalia c-Opsin.
To address the question of whether the identified Tripedalia c-opsin can function as a true visual opsin, we tested its photochemical properties. Tripedalia c-opsin was expressed in COS-1 cells and reconstituted as a functional photosensitive pigment with 11-cis- retinal. The reconstituted c-opsin was most sensitive to the blue–green region of the spectrum with a peak absorbance (λmax) at 465–470 nm (Fig. 1H), in agreement with the spectral sensitivity of the Tripedalia electroretinogram (16). We conclude that Tripedalia c-opsin is a functional, vertebrate-like photopigment expressed in the PRCs of the camera-type cubozoan eye.
Tripedalia Orthologues of Vertebrate-Like Phototransduction Genes.
In vertebrates, activated heterotrimeric G proteins use cGMP PDE for signal transduction. In accordance with our identification of a vertebrate-like c-opsin in Tripedalia, we found that the catalytic subunit of pde expressed in the Tripedalia rhopalium phylogenetically clusters with the group of GAF domain-containing PDEs including vertebrate rod- and cone-specific PDE6 (Fig. S2A). Furthermore, we identified other components of the ciliary-type cascade associated with deactivation or adaptation of phototransduction, such as the inhibitory subunit of phosphodiesterase (PDE6D), phosducin and guanylate cyclase (Fig. S2 B–D). Thus, the nature of the genes expressed in the rhopalia (detected by RT-PCR; Fig. S3) suggests that the camera-type eye of Tripedalia uses a ciliary-type phototransduction cascade similar to that of vertebrates.
Melanin Granules in Tripedalia PRCs.
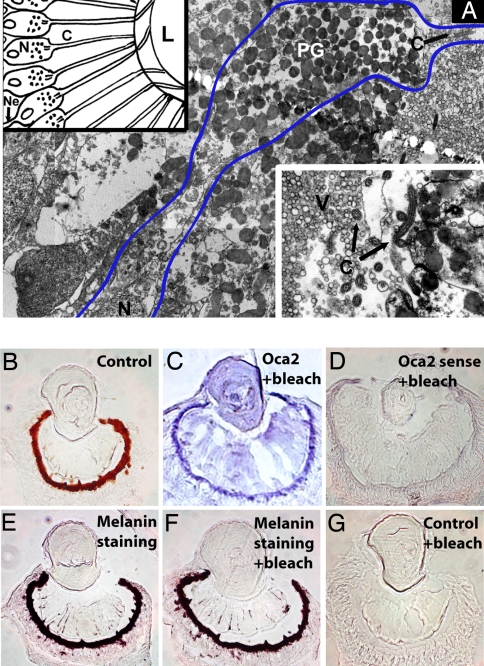
A conspicuous ring of dark shielding pigment surrounds the area of c-opsin expression (compare Fig. 1 D and E). Most if not all PRCs in the Tripedalia retina contain pigment granules (Fig. 2A). These PRCs thus resemble what one might imagine a prototypical ancestral cell combining photoreceptor and pigment functions to look like (17). The biochemical nature of the Tripedalia pigment was suggested by the identification of an orthologue of the vertebrate ocular and cutaneous albinism-2 (Oca2) gene in our EST library (Fig. S4A). Oca2 (also known as pink-eyed dilution) is an essential gene for melanin biosynthesis. It is the most commonly mutated gene in cases of human albinism (18). In addition, mutations in Oca2 are responsible for pigmentation defects in mouse (19), medaka (20), and independently arisen populations of the cave fish, Astyanax (21). In situ hybridization analysis revealed that Tripedalia oca is conspicuously expressed near the pigmented retina layer of PRCs (Fig. 2C). Control staining with an oca sense probe (Fig. 2D) did not yield a signal. The results of a direct chemical assay (Fontana–Masson) were consistent with melanin being the pigment in the Tripedalia retina (Fig. 2 E–G). In vertebrates, development of melanin-producing cells and specification of retinal pigment cells require the conserved microophthalmia-associated transcription factor, Mitf (22–24). Mitf regulates expression of tyrosinase and tyrosinase-related protein-1 and -2, which are necessary for melanin biosynthesis (for review, see ref. 25). Here, we have cloned a mitf orthologue from Tripedalia (Fig. S4B) expressed in a ring-like pattern just outside of the melanin deposits (Fig. 3 A and B); this area contains the PRC nuclei (Figs. 2A and 3D). Thus, mitf is a conserved transcription factor with shared expression patterns in the complex eyes of Tripedalia and vertebrates.
Fig. 2.
Melanin is the shielding pigment of Tripedalia camera-type eyes. (A) Electron micrograph of camera-type eye PRCs. Blue line borders one PRC containing shielding pigment layer (PG) as well as photosensitive cilium (C). (Upper Left Inset) Diagram of PRCs with their cilia protruding to the lens capsule. Note that all PRC nuclei are located behind the shielding pigment. (Lower Right Inset) Apical end of a PRC with cilium. C, cilium; L, lens; N, nucleus; Ne, neurite; PG, pigment granules; V, vitreous body. (B) Bright-field cryosection of camera-type eye. (C and D) In situ hybridization (blue) using oca2 antisense (C) or control sense (D) probes. (E–G) Melanin detection by Fontana–Masson staining. Tissue sections shown in C, D, F, and G were bleached to remove melanin.
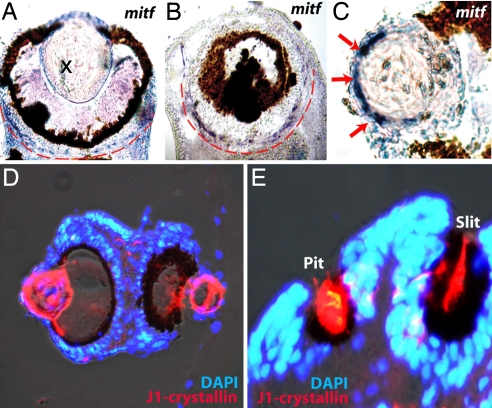
Fig. 3.
Expression of mitf and J1-crystallin in Tripedalia eyes. (A–C) In situ hybridization (blue) detects mitf expression in the circle around pigment deposits (A and B) and in the lens (C, arrows). (D and E) Immunohistochemistry staining using an antibody to the major cubozoan lens crystallin, J1 (red). Nuclei of cells are visualized by DAPI staining (blue). J1-crystallin expression is localized to lenses of camera-type eyes (D) as well as to the slit and pit eyes (E).
Mitf Expression in Lens and Crystallin Expression in Pigmented PRCs of Nonlens Eyes of Tripedalia.
In addition to expression in the pigmented PRCs of camera-type eyes, mitf mRNA was detected in the outermost cells of the Tripedalia lens (Fig. 3C). Consequently, to investigate a possible relationship between the pigmented PRCs and the cellular lens we examined whether J1-crystallin, the major protein of the Tripedalia lens (26), is expressed in PRCs. The J1-crystallin antibody immunostained the slit and pit eyes as well as the cellular lens (Fig. 3 D and E). The presence of J1-crystallin in the slit and pit eyes of Tripedalia was unexpected because these cup-like eyes lack cellular lenses. They do, however, have pigmented PRCs, suggesting a relationship between the PRCs and cellular lens that warrants further study.
Discussion
The present work reveals surprising similarities in the genetic components used for visual system development in vertebrates and cubozoan jellyfish. If Cubozoa and vertebrates express orthologous c-opsins in their PRCs and make use of the same pigmentation pathway including the key transcription factor Mitf, does this represent a parallel evolution or conservation of an ancestral “eye” program between those evolutionarily distant animal phyla (Fig. 4)? Although our data are formally consistent with both evolutionary scenarios, we believe that they favor the former.
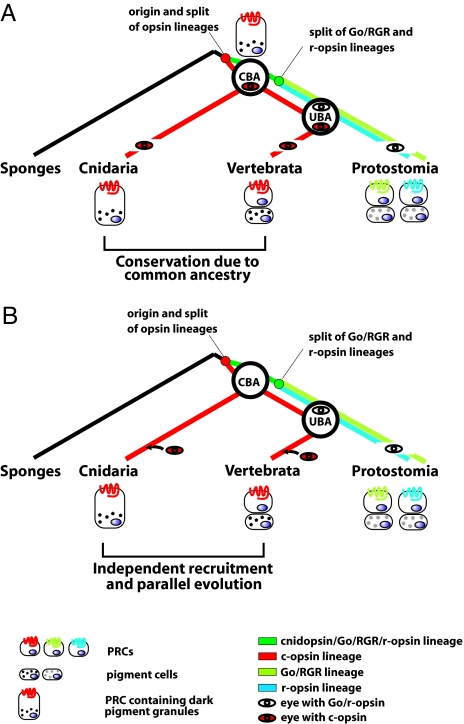
Fig. 4.
Two scenarios for the use of ciliary phototransduction and melanogenic pathway in eye evolutionary history. A simplified view of the two evolutionary scenarios is compatible with the data in the present work. The use of similar genetic components in vertebrate and cubozoan eyes is either due to common ancestry (A) or independent parallel recruitments in cnidarian and vertebrate lineages (B). The c-opsins and Go/r-opsins arose by duplication and diversification of an ancestral opsin in the early metazoans (27). In the schemes, only the visual (i.e., the eye-specific) PRCs and opsins are considered. Different shading of pigment granules indicates possible distinct chemical composition. CBA, cnidarian–bilaterian ancestor; UBA, urbilaterian ancestor.
Even though ciliary and rhabdomeric photoreceptive systems coexist throughout the animal taxa (1), the present evidence suggests that their evolutionary histories differ. For photodetection, all invertebrate PRCs examined employ Go/r-opsin, and all vertebrate PRCs employ c-opsin (2, 3). Importantly, c-opsin is expressed in the ciliary PRCs in the brain of the polychaete worm, P. dumerili, whereas r-opsin is expressed in rhabdomeric PRCs in the eyes (13). Based on this result, Arendt et al. (13) have proposed that early metazoans possessed a single type of PRC with an ancestral opsin for light detection that later diversified into two distinct PRC and opsin types. The rhabdomeric PRCs (with r-opsin) were used in the eyes for photoreception, whereas the ciliary PRCs (with c-opsin) were incorporated into the evolving brain. These findings are consistent with vertebrates confining r-opsin to retinal ganglion cells apparently for photoperiodicity and using ciliary PRCs containing c-opsins exclusively for photoreception (rods and cones). Taken together, the data suggest that the ganglion cells of the vertebrate retina are the evolutionary descendents of rhabdomeric PRCs (3, 13). Moreover, because no identified opsin gene in cnidarians (27) contains a typical r-opsin fingerprint tripeptide HPK critical for coupling to the downstream phototransduction cascade, it was proposed that r-opsins are a bilaterian innovation that originated after the separation of the cnidarian and bilaterian lineages (Fig. 4) (27).
All eyes have shielding pigment typically found in cells adjacent to the PRCs. Melanin, the dark pigment of Tripedalia eyes, presumably performs the same function in vertebrate eyes as in the simple cup-like eyes of a basal lophotrochozoan, Dugesia (6). Interestingly, Dugesia uses another pigment, an ommochrome, as the body pigment (2, 6). Pterins constitute the dark eye pigment of the polychaete P. dumerilii (4), and pterins and ommochromes are the pigments in eyes of Drosophila (5). Thus, as with the opsins, Tripedalia shares the same dark pigment in the eye with vertebrates.
Unlike in the Dugesia eye, the camera-type Tripedalia eye combines the photoreceptor and pigment functions in the same cell consistent with an ancestral (basal) condition (Fig. 4A). However, the medusae stage of cubozoans may well be a derived rather than ancestral condition for Cnidaria, complicating discussions about the basal state of the cubozoan visual system (Anthozoans, for example, do not have eyes). Nevertheless, it remains possible that the pigmented PRCs in Tripedalia are descendants of one of the postulated ancient prototypical photosensitive cells diversified by natural selection (17, 28). However, this does not require a common origin for the eyes. It was estimated through computer-based modeling (29) that fewer than a half-million generations would be required under selective pressure to proceed from a cluster of light-sensitive cells to a sophisticated camera-type eye. In theory, this relatively short time interval would allow sophisticated eyes to have originated de novo several times during evolution (polyphyletic eye origin).
For the common-ancestry model to be true, the cnidarian-bilaterian ancestor (CBA) must have had the same genetic determinants as its descendants. The common-ancestry scenario for cubozoan and vertebrate eyes requires, however, that animals in many bilaterian phyla lost their eyes that were initially assembled by using the same building blocks as in present-day vertebrates and Cubozoa (c-opsins, melanin) to explain the exclusive occurrence of rhabdomeric PRCs in invertebrate eyes. There is no obvious explanation for such a specific selection against ciliary PRCs to be used for visual purposes. Eyes in general provide a freely moving animal with a tremendous advantage, and as such there should be a constant selection for eye maintenance, except in, for example, cave or underground animals.
Although not definitive, there are at least two additional complications to the common-ancestry model that arise if one invokes the developmental argument that similar transcription factor cascades may direct development of vertebrate and cubozoan eyes. The first is that PaxB, a Pax2/6/8-related transcription factor, is used in Tripedalia (30) instead of Pax6 as in vertebrates (31) as well as flies (32, 33) and other species (34). The second is the apparent evolutionary “promiscuity” of developmental cascades in general; entire regulatory circuits can be co-opted for development of different cell types, tissues, or organs. For example, the Pax–Six–Eya–Dach gene regulatory network has a fundamental role in Drosophila visual system development but is also used for specification of muscle cells or placodes in vertebrates (35). Co-opting orthologous suites of genes for similar functions could be a possible explanation for independent or parallel evolution of cubozoan and vertebrate eyes with ciliary-type PRCs (Fig. 4B) (1, 36, 37). Independent derivation of Tripedalia and vertebrate eyes would also fit conceptually with the early idea that PRCs originated multiple times (38), although it does not address how many times PRCs themselves may have originated. That vertebrates and Cnidaria share many more genes than anticipated (39, 40), including pax, mitf, c-opsin, pde's, phosducin, guanylate cyclase, and oca2 (ref. 30 and this work), supports the notion that both animal groups use similar sets of genes to generate significantly different body plans. It follows that changes in gene regulation, rather than “new” genes, may drive novelties such as eyes during evolution. Finally, ectopic eye formation by misexpression of Pax6 provides an astounding example of how an eye might arise de novo in a foreign tissue environment (33, 34). The fact that ectopic eyes can be generated experimentally suggests that the same gene, Pax, used by various eyes of present-day animals could have been instrumental in creating eyes independently numerous times during evolution.
In addition to sharing the same genetic building blocks in their PRCs (ciliary phototransduction, melanogenic pathway), cubozoans and vertebrates both use a cellular lens to increase visual sensitivity and produce a sharp image in the desired plane of focus. The optical properties of cellular lenses are caused by the high-level expression of proteins collectively called crystallins (ref. 41 and this work). In striking contrast to the conservation of opsins as the visual pigments in the PRCs, the lens crystallins are diverse proteins that are often taxon-specific, i.e., entirely different proteins function as crystallins in different species. Similar transcription factors including those of the Pax gene family have been independently recruited for the regulation of nonhomologous crystallin genes in Tripedalia and vertebrates (30, 42, 43) to achieve a gradient of refractive index within their transparent lenses. The independent recruitment of lens crystallins is consistent with parallel evolution of cubozoan and vertebrate eyes and provides a striking example of the role of convergence in eye evolution.
Finally, the present findings of mitf in the lens and J1-crystallin in the pigmented slit and pit ocelli of Tripedalia support the idea that the cellular cubozoan lens arose from a pigmented cell ancestor. It is known that pigment cells may acquire the capacity to secrete lens-forming material (44). Combined, our data on J1-crystallin and mitf expression suggest that the cellular cubozoan lens with its remarkable ability to refract light without spherical aberration (11) originated from a pigment cell ancestor and that the primitive cup-like eyes located on the cubozoan rhopalia might be evolutionary forerunners of camera-type eyes.
In conclusion, the present study uncovers a surprising molecular parallelism in the eye design of vertebrates and cubozoan jellyfish. Although the current data do not distinguish unambiguously between the common-ancestry and independent-recruitment scenarios, we propose that they lean in the direction of the latter, favoring multiple independent reorganizations of common elements and independent recruitments of similar suites of genes during evolution of the diverse eyes.
Materials and Methods
Jellyfish Collection and Culture.
T. cystophor a was collected and cultured as described in ref. 43.
Isolation of Rhopalium-Expressed Genes and Phylogenetic Analysis.
An EST cDNA library was generated from rhopalia mRNA, and 2,433 individual clones from the library were sequenced by using an ABI capillary sequencer. The accession numbers for the clones are as follows: c-opsin (EU310498), oca (EU310502), mitf (EU310499), catalytic pde (EU310500), inhibitory pde6d (EU310501) and guanylate cyclase (EU310503). Details on phylogenetic analysis including the accession numbers of individual sequences are described in SI Materials and Methods.
RNA in Situ Hybridization.
Jellyfish were fixed in 4% paraformaldehyde (PFA), cryoprotected in 30% sucrose overnight at 4°C, and embedded and frozen in OCT (Tissue Tek). RNA in situ hybridization was performed as described in ref. 43.
Immunohistochemistry.
The cryosections were refixed in 4% PFA for 10 min, washed three times with PBS, permeabilized with PBT (PBS + 0.1% Tween 20) for 15 min, and blocked in 10% BSA in PBT for 30 min. The primary antibodies were diluted in 1% BSA in PBT, incubated overnight at room temperature, washed three times with PBS, and incubated with secondary antibodies in 1% BSA in PBT. The sections were counterstained with DAPI and mounted. Primary antibodies used were: anti-Tripedalia c-opsin, anti-Tripedalia J1-crystallin, and anti-acetylated tubulin (Sigma). The following secondary antibodies were used: Alexa Fluor 488- or 594-conjugated goat anti-mouse or anti-rabbit IgG (Molecular Probes).
Generation of Antibodies, COS-7 Cell Transfection, and Western Blotting.
Antibodies directed against Tripedalia c-opsin and J1-crystallin were prepared by immunization of rabbits as follows. The C-terminal region of c-opsin cDNA corresponding to amino acids 274–329 was cloned into the expression vector pET42, expressed in BL21(DE3)RIPL cells (Stratagene), and purified by using His6 tag chromatography. The N-terminal peptide of J1-crystallin AAIVGSLVADAATQPVHK was attached to KLH via the C-terminal lysine and used for immunization. Monkey kidney COS-7 cells were transfected with CMV-c-opsin (amino acids 1–329) expression vector by using FuGENE6 reagent (Roche). Total extracts were prepared from c-opsin-transfected cells, mock-transfected cells, and rhopalia and were analyzed by Western blotting by using anti-c-opsin rabbit serum and chemiluminescent detection kit (Pierce). To avoid formation of multimeric opsin complexes, protein extracts from transfected cells were diluted and heated at low temperature (37°C) before SDS/PAGE.
Fontana–Masson Method.
The cryosections were hydrated in distilled water and then incubated with Fontana silver nitrate working solution (2.5% silver nitrate) at 56°C for 1–2 h. After three washes in distilled water, sections were treated in 0.2% gold chloride at room temperature for 2 min, rinsed once in distilled water, placed in 5% sodium thiosulfate at room temperature for 1 min, washed again in water, and mounted.
Melanin Bleach Procedure.
Bleaching was performed either after Fontana–Masson staining or RNA in situ hybridization. The sections were hydrated in distilled water and exposed to 0.25% potassium permanganate for 30 min at room temperature. The sections were treated with 5% oxalic acid for 5 min, washed with water, and mounted.
Transmission Electron Microscopy.
Rhopalia excised from juvenile medusae were treated with Karnovsky fixative (2.5% glutaraldehyde, 2.5% paraformaldehyde in cacodylate buffer) for 24 h at 4°C. Fixed tissue was washed 12 h in 0.1% cacodylate buffer at 4°C. Karnovsky-fixed juvenile rhopalia and PFA-fixed adult rhopalia were postfixed in 2% OsO4 for 2 h at 4°C and then washed in water. Samples were dehydrated in series of ethanol solutions, transferred to pure acetone, and embedded in Poly/Bed 812/Araldite 502 resin. Ultrathin sections (600–800 nm) were cut on Ultracut E (Reichert–Jung), placed on copper grids, and treated with 2.5% uranyl acetate for 1 h followed by lead citrate for 15 min. The material was examined by transmission electron microscopy (Jeol-1011), and images were taken with a MEGAview III Soft imaging system.
Expression, Reconstitution, and Spectroscopic Analysis of Tripedalia c-opsin.
Tripedalia c-opsin cDNA was expressed in transfected COS-1 cells. Transfected cells were resuspended with 5 μM 11-cis-retinal, solubilized with 1% dodecyl maltoside, and the resulting c-opsin photopigment was purified by using immobilized 1D4 (Cell Culture Center, Minneapolis, MN). The UV-visible absorption spectrum was recorded for the c-opsin photopigment from 250 to 650 nm at 0.5-nm intervals by using the Hitachi U3010 dual-beam spectrometer at 20°C. Five replicates were performed in the dark and five more after 3 min of light exposure (with a <440-nm cut-off filter). The λmax value was taken from the dark–light difference spectrum.
For additional details, see SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Drs. Ales Cvekl and Stanislav Tomarev for comments on the manuscript and Mrs. Veronika Noskova for excellent technical assistance. We are grateful to Prof. Tom Tosteson for kind support during our collecting trip to Puerto Rico. This work was supported in part by Project AV0Z50520514 awarded by the Academy of Sciences of the Czech Republic and by Center for Applied Genomics Grant 1M6837805002 awarded by the Ministry of Education, Youth, and Sports of the Czech Republic and by the intramural research program of the National Eye Institute, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. EU310498 (c-opsin), EU310502 (oca), EU310499 (mitf), EU310500 (catalytic pde), EU310501 (inhibitory pde6d) and EU310503 (guanylate cyclase)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0800388105/DCSupplemental.
References
- 1.Fernald RD. Casting a genetic light on the evolution of eyes. Science. 2006;313:1914–1918. doi: 10.1126/science.1127889. [DOI] [PubMed] [Google Scholar]
- 2.Arendt D, Wittbrodt J. Reconstructing the eyes of Urbilateria. Philos Trans R Soc London Ser B. 2001;356:1545–1563. doi: 10.1098/rstb.2001.0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arendt D. Evolution of eyes and photoreceptor cell types. Int J Dev Biol. 2003;47:563–571. [PubMed] [Google Scholar]
- 4.Viscontini M, Hummel W, Fischer A. Isolation of pterin dimers from the eyes of Platynereis dumerilii (German) Helv Chim Acta. 1970;53:1207–1209. [Google Scholar]
- 5.Shoup JR. The development of pigment granules in the eyes of wild-type and mutant Drosophila melanogaster. J Cell Biol. 1966;29:223–249. doi: 10.1083/jcb.29.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hase S, et al. Characterization of the pigment produced by the planarian, Dugesia ryukyuensis. Pigment Cell Res. 2006;19:248–249. doi: 10.1111/j.1600-0749.2006.00306.x. [DOI] [PubMed] [Google Scholar]
- 7.Conant FS. Notes on the Cubomedusae. Johns Hopkins Univ Circ. 1897;132:8–10. [Google Scholar]
- 8.Pearse JS, Pearse VB. Vision of cubomedusan jellyfishes. Science. 1978;199:458. doi: 10.1126/science.22934. [DOI] [PubMed] [Google Scholar]
- 9.Yamasu T, Yoshida M. Fine structure of complex ocelli of a cubomedusan, Tamoya bursaria. Haeckel Cell Tissue Res. 1976;170:325–339. doi: 10.1007/BF00219415. [DOI] [PubMed] [Google Scholar]
- 10.Garm A, O'Connor M, Parkefelt L, Nilsson DE. Visually guided obstacle avoidance in the box jellyfish Tripedalia cystophora and Chiropsella bronzie. J Exp Biol. 2007;210:3616–3623. doi: 10.1242/jeb.004044. [DOI] [PubMed] [Google Scholar]
- 11.Nilsson DE, Gislen L, Coates MM, Skogh C, Garm A. Advanced optics in a jellyfish eye. Nature. 2005;435:201–205. doi: 10.1038/nature03484. [DOI] [PubMed] [Google Scholar]
- 12.Terakita A. The opsins. Genome Biol. 2005;6:213. doi: 10.1186/gb-2005-6-3-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arendt D, Tessmar-Raible K, Snyman H, Dorresteijn AW, Wittbrodt J. Ciliary photoreceptors with a vertebrate-type opsin in an invertebrate brain. Science. 2004;306:869–871. doi: 10.1126/science.1099955. [DOI] [PubMed] [Google Scholar]
- 14.Raible F, et al. Opsins and clusters of sensory G protein-coupled receptors in the sea urchin genome. Dev Biol. 2006;300:461–475. doi: 10.1016/j.ydbio.2006.08.070. [DOI] [PubMed] [Google Scholar]
- 15.Marin EP, et al. The amino terminus of the fourth cytoplasmic loop of rhodopsin modulates rhodopsin-transducin interaction. J Biol Chem. 2000;275:1930–1936. doi: 10.1074/jbc.275.3.1930. [DOI] [PubMed] [Google Scholar]
- 16.Coates MM, Garm A, Theobald JC, Thompson SH, Nilsson DE. The spectral sensitivity of the lens eyes of a box jellyfish, Tripedalia cystophora (Conant) J Exp Biol. 2006;209:3758–3765. doi: 10.1242/jeb.02431. [DOI] [PubMed] [Google Scholar]
- 17.Arnheiter H. Evolutionary biology: Eyes viewed from the skin. Nature. 1998;391:632–633. doi: 10.1038/35487. [DOI] [PubMed] [Google Scholar]
- 18.Oetting WS, Garrett SS, Brott M, King RA. P gene mutations associated with oculocutaneous albinism type II (OCA2) Hum Mutat. 2005;25:323. doi: 10.1002/humu.9318. [DOI] [PubMed] [Google Scholar]
- 19.Rinchik EM, et al. A gene for the mouse pink-eyed dilution locus and for human type II oculocutaneous albinism. Nature. 1993;361:72–76. doi: 10.1038/361072a0. [DOI] [PubMed] [Google Scholar]
- 20.Fukamachi S, et al. Conserved function of medaka pink-eyed dilution in melanin synthesis and its divergent transcriptional regulation in gonads among vertebrates. Genetics. 2004;168:1519–1527. doi: 10.1534/genetics.104.030494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Protas ME, et al. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat Genet. 2006;38:107–111. doi: 10.1038/ng1700. [DOI] [PubMed] [Google Scholar]
- 22.Hodgkinson CA, et al. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix–loop–helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen M, Arnheiter H. Signaling and transcriptional regulation in early mammalian eye development: A link between FGF and MITF. Development. 2000;127:3581–3591. doi: 10.1242/dev.127.16.3581. [DOI] [PubMed] [Google Scholar]
- 24.Opdecamp K, et al. Melanocyte development in vivo and in neural crest cell cultures: Crucial dependence on the Mitf basic helix–loop–helix zipper transcription factor. Development. 1997;124:2377–2386. doi: 10.1242/dev.124.12.2377. [DOI] [PubMed] [Google Scholar]
- 25.Goding CR. Mitf from neural crest to melanoma: Signal transduction and transcription in the melanocyte lineage. Genes Dev. 2000;14:1712–1728. [PubMed] [Google Scholar]
- 26.Piatigorsky J, Horwitz J, Norman BL. J1-crystallins of the cubomedusan jellyfish lens constitute a novel family encoded in at least three intronless genes. J Biol Chem. 1993;268:11894–11901. [PubMed] [Google Scholar]
- 27.Plachetzki DC, Degnan BM, Oakley TH. The origins of novel protein interactions during animal opsin evolution. PLoS ONE. 2007;2:e1054. doi: 10.1371/journal.pone.0001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gehring WJ, Ikeo K. Pax 6: Mastering eye morphogenesis and eye evolution. Trends Genet. 1999;15:371–377. doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
- 29.Nilsson DE, Pelger S. A pessimistic estimate of the time required for an eye to evolve. Proc Biol Sci. 1994;256:53–58. doi: 10.1098/rspb.1994.0048. [DOI] [PubMed] [Google Scholar]
- 30.Kozmik Z, et al. Role of Pax genes in eye evolution: A cnidarian PaxB gene uniting Pax2 and Pax6 functions. Dev Cell. 2003;5:773–785. doi: 10.1016/s1534-5807(03)00325-3. [DOI] [PubMed] [Google Scholar]
- 31.Hill RE, et al. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- 32.Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the small eye gene in mice and Aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- 33.Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- 34.Gehring WJ. Historical perspective on the development and evolution of eyes and photoreceptors. Int J Dev Biol. 2004;48:707–717. doi: 10.1387/ijdb.041900wg. [DOI] [PubMed] [Google Scholar]
- 35.Silver SJ, Rebay I. Signaling circuitries in development: Insights from the retinal determination gene network. Development. 2005;132:3–13. doi: 10.1242/dev.01539. [DOI] [PubMed] [Google Scholar]
- 36.Fernald RD. Eyes: Variety, development, and evolution. Brain Behav Evol. 2004;64:141–147. doi: 10.1159/000079743. [DOI] [PubMed] [Google Scholar]
- 37.Fernald RD. Evolving eyes. Int J Dev Biol. 2004;48:701–705. doi: 10.1387/ijdb.041888rf. [DOI] [PubMed] [Google Scholar]
- 38.Salvini-Plawen LV, Mayr E. On the evolution of photoreceptors and eyes. Evol Biol. 1977;10:207–263. [Google Scholar]
- 39.Kortschak RD, Samuel G, Saint R, Miller DJ. EST analysis of the cnidarian Acropora millepora reveals extensive gene loss and rapid sequence divergence in the model invertebrates. Curr Biol. 2003;13:2190–2195. doi: 10.1016/j.cub.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 40.Putnam NH, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 41.Piatigorsky J. Gene Sharing and Evolution: The Diversity of Protein Functions. Cambridge, MA: Harvard Univ Press; 2007. [Google Scholar]
- 42.Cvekl A, Yang Y, Chauhan BK, Cveklova K. Regulation of gene expression by Pax6 in ocular cells: A case of tissue-preferred expression of crystallins in lens. Int J Dev Biol. 2004;48:829–844. doi: 10.1387/ijdb.041866ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kozmik Z, et al. Cubozoan crystallins: Evidence for convergent evolution of Pax regulatory sequences. Evolution Dev. 2008;10:52–61. doi: 10.1111/j.1525-142X.2007.00213.x. [DOI] [PubMed] [Google Scholar]
- 44.Eakin RM, Westfall JA. Further observation on the fine structure of some invertebrate eyes. Z Zellforsch Mikrosk Anat. 1964;62:310–332. doi: 10.1007/BF00339283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.