Abstract
The catalytic domain of protein kinases harbors a large number of disease-causing single nucleotide polymorphisms (SNPs) and common or neutral SNPs that are not known or hypothesized to be associated with any disease. Distinguishing these two types of polymorphisms is critical in accurately predicting the causative role of SNPs in both candidate gene and genome-wide association studies. In this study, we have analyzed the structural location of common and disease-associated SNPs in the catalytic domain of protein kinases and find that, although common SNPs are randomly distributed within the catalytic core, known disease SNPs consistently map to regulatory and substrate binding regions. In particular, a buried side-chain network that anchors the flexible activation loop to the catalytic core is frequently mutated in disease patients. This network was recently shown to be absent in distantly related eukaryotic-like kinases, which lack an exaggerated activation loop and, presumably, are not regulated by phosphorylation.
Keywords: allostery, cancer, conservation, evolution, mutation
Protein kinases are a large family of evolutionarily related proteins that control numerous signaling pathways in the eukaryotic cell. They share a conserved catalytic core, which catalyzes the transfer of the γ-phosphate from ATP to the hydroxyl group of serine, threonine or tyrosine in protein substrates (1). Addition of this phosphate moiety can modulate enzyme activity, it can serve as a docking site for other proteins, or it can exert allosteric regulatory effects. Because many fundamental cellular processes, such as transcription, translation, and cytoskeletal reorganization, are regulated by protein phosphorylation, the catalytic activity of protein kinases involved in these pathways is very tightly controlled. Abnormal activation or regulation of protein kinases is a major cause of human disease (2, 3), especially cancers and malformation syndromes (4, 5). Although several factors can cause misregulation of proteins, missense mutations (also referred to as nsSNPs) remain one of the main causes (6).
Historically, emphasis has been placed on protein coding variations [i.e., nonsynonymous coding SNPs (nsSNPs)] in mediating disease susceptibility, and several such variations, typically rare, have been unequivocally shown to influence disease susceptibility, especially in the context of overt Mendelian disorders (7). It is estimated that 67,000–200,000 nsSNPs occur naturally in the human population at large (8). However, both the overall degree to which nsSNPs influence disease and the frequency of these nsSNPs are unknown. As a result, some researchers have turned to Whole Genome Association (WGA) studies and large-scale studies of nsSNPs to find DNA sequence variations that influence diseases.
Although quite powerful, such large-scale studies are hampered by potential heterogeneity of the disease in question, gene-by-environment interactions, and multiple testing issues (9). A possible solution to these problems is to computationally prioritize candidate nsSNPs to be tested for association with a disease. A few methods have been designed for this purpose (10, 11). Structural information from representative solved crystal structures of a particular gene family can be used to derive sequence-based properties of a large collection of variations. In this way, researchers can gain insight into the functional significance of particular nsSNPs and the functional significance of key residues within a specific protein family.
Here, we focus on the protein kinase gene family, the catalytic domain of which was recently shown to harbor a large number of rare (frequency <1%) single nucleotide polymorphisms (SNPs) that underlie inherited disease (12). The catalytic domain, however, also harbors common SNPs (frequency >1%), the majority of which are not thought to cause disease (12). Although both common and disease-causing SNPs occur outside of the catalytic domain, structural homology across all kinases allows for an examination of the sequence-based and structural properties of the disease-causing vs. non-disease-causing nsSNPs within the catalytic domain and may reveal important biomedical features of kinases and help make sense of variations either targeted or merely identified in genetic association studies. To this end, we systematically catalogued disease and common SNPs (12) residing within the kinase catalytic core and then mapped them to individual subdomains, which are characterized by patterns of conserved residues and whose functions are known to varying degrees (13). Rigorous statistical methods were then used to identify residue positions that are significantly overrepresented among disease vs. common SNPs. Mapping of these germ-line SNPs associated primarily with developmental and metabolic disorders will enable more accurate predictions of common SNP functionality and accurate evaluation of the affect of mutations in the kinome of various cancers (14). Note that we refer to common SNPs not known to cause disease as “common SNPs” and nonsynonymous coding SNPs (nsSNPs) as “SNPs” for brevity.
Surprisingly, our analyses suggest that a significant number of disease-associated nsSNPs are not directly involved in ATP binding or catalysis but rather are buried in the catalytic core. Structural analysis of these residues suggests that they are frequently involved in substrate binding and regulation. In particular, a conserved side-chain network, which anchors the flexible activation loop to the catalytic core, is profoundly affected in many human disease states. This result could not have been anticipated or appreciated without an in-depth study of the unique evolutionary and functional features of kinases.
Results
Distribution of Disease-Causing vs. Common SNPs Within the Catalytic Core.
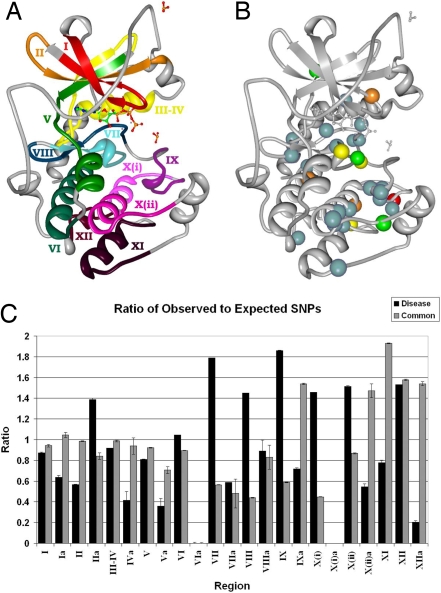
To determine the distribution of disease and common SNPs within the catalytic domain, we represented the catalytic domain by 12 characteristic subdomains as defined by Hanks and Hunter (13, 15) and by intervening regions connecting these subdomains [Fig. 1A and supporting information (SI) Table S1]. Mapping of common and disease SNPs to these regions (described in Methods) revealed strikingly different distributions (Fig. 1C and Table 1). Specifically, the distribution of common SNPs within subdomains and intervening regions conforms to random or chance expectations, whereas disease SNPs tend to occur more frequently than expected within subdomains and less frequently within intervening regions (P = 0.0006). Statistical significance is evaluated by using the binomial distribution, where the probabilities are adjusted for the length of each subdomain (see Methods for further details). To verify that the difference in these distributions is not a result of bias in subdomain length, we compared the average lengths of corresponding regions across the proteins containing common or disease SNPs and observed no significant differences (P = 0.8269). Thus, although both disease and common SNPs are widely distributed throughout the catalytic core, they occur with different frequencies within subdomains and intervening regions.
Fig. 1.
Kinase subdomains and SNP distribution. (A) The subdomains PKA (PDB entry 1ATP). Gray residues are intervening loops. Subdomains are numbered by roman numerals and color coded. (B) The distribution of kinase disease SNPs. Spheres denote residues with high disease SNP frequencies; red, eight SNPs; yellow, seven SNPs; orange, six SNPs; green, five SNPs; and blue, four SNPs. (C) Ratio of observed to expected SNPs per region. Roman numerals correspond to subdomains in A, where a denotes the intervening region between subdomains. Black bars, disease SNPs; gray bars, common SNPs. The image was created in part with Protein Workshop (54).
Table 1.
Subdomain Distribution of SNPs
| Subdomain | Common |
Disease |
Comparison |
|||||
|---|---|---|---|---|---|---|---|---|
| Length, % | SNPs, % | P | Length, % | SNPs, % | P | Length | SNPs | |
| I (43–60) | 5.98 | 5.63 | 0.8965 | 6.16 | 5.37 | 0.4592 | 0.9704 | 0.9091 |
| Ia (61–62) | 1.35 | 1.41 | 1.0000 | 1.46 | 0.93 | 0.4471 | 0.9628 | 0.6604 |
| II (63–77) | 5.14 | 5.07 | 1.0000 | 5.36 | 3.04 | 0.0209* | 0.9616 | 0.3049 |
| Iia (78–84) | 2.34 | 1.97 | 0.8160 | 1.69 | 2.34 | 0.4365 | 0.8179 | 0.8010 |
| III-IV (85–114) | 10.55 | 10.42 | 1.0000 | 10.70 | 9.81 | 0.4582 | 0.9817 | 0.8398 |
| IVa (115) | 2.10 | 1.97 | 1.0000 | 1.69 | 0.70 | 0.1171 | 0.8826 | 0.2631 |
| V (116–134) | 6.42 | 5.92 | 0.8050 | 6.62 | 5.37 | 0.2609 | 0.9681 | 0.8145 |
| Va (135–138) | 2.39 | 1.69 | 0.5083 | 5.23 | 1.87 | 0.0004* | 0.5009 | 0.8919 |
| VI (139–159) | 7.23 | 6.48 | 0.6729 | 7.36 | 7.71 | 0.9973 | 0.9807 | 0.6313 |
| VIa (−) | 0.17 | 0.00 | 1.0000 | 0.56 | 0.00 | 0.1635 | 0.7663 | 1.0000 |
| VII (160–175) | 5.49 | 3.10 | 0.0487* | 5.61 | 10.05 | 0.0008* | 0.9798 | 0.0031* |
| VIIa (176) | 2.92 | 1.41 | 0.1032 | 0.40 | 0.23 | 0.9463 | 0.2184 | 0.1701 |
| VIII (177–191) | 5.13 | 2.25 | 0.0107* | 5.32 | 7.71 | 0.0680 | 0.9671 | 0.0079* |
| VIIIa (192–198) | 5.08 | 4.23 | 0.5530 | 4.72 | 4.21 | 0.6055 | 0.9355 | 0.9921 |
| IX (199–212) | 4.78 | 2.82 | 0.0923 | 4.90 | 9.11 | 0.0007* | 0.9786 | 0.0050* |
| IXa (213–214) | 1.10 | 1.69 | 0.3962 | 1.30 | 0.93 | 0.6335 | 0.9301 | 0.5074 |
| X(i) (215–225) | 3.77 | 1.69 | 0.0381* | 3.85 | 5.61 | 0.1213 | 0.9846 | 0.0276* |
| X(i)a (-) | 0.02 | 0.00 | 1.0000 | <0.0001 | 0.00 | 1.0000 | 0.9033 | 1.0000 |
| X(ii) (226–240) | 5.19 | 4.51 | 0.6651 | 5.87 | 8.88 | 0.0267* | 0.8884 | 0.0751 |
| X(ii)a (241–256) | 9.76 | 14.37 | 0.0071* | 7.26 | 3.97 | 0.0038* | 0.6614 | 0.0002* |
| XI (257–279) | 7.88 | 15.21 | <0.0001* | 8.12 | 6.31 | 0.1313 | 0.9662 | 0.0036* |
| XII (280–294) | 3.57 | 5.63 | 0.0639 | 3.51 | 5.37 | 0.0850 | 0.9873 | 0.9091 |
| XIIa (−) | 1.65 | 2.54 | 0.2706 | 2.30 | 0.47 | 0.0043* | 0.8261 | 0.0747 |
| Sub-domains | 71.12 | 68.73 | 0.3320 | 73.37 | 84.35 | <0.0001* | 0.8269 | 0.0006* |
| Intervening | 28.88 | 31.27 | 0.3320 | 26.62 | 15.65 | <0.0001* | 0.8269 | 0.0006* |
Subdomains are identified by Roman numeral numbering, and PKA positions are in parentheses.
*Statistically significant.
The Substrate Binding C-Lobe of the Kinase Core Is Enriched in Disease SNPs.
We next examined the distribution of common and disease SNPs within the individual subdomains of the catalytic core. The ratio of expected to observed SNPs is shown in Fig. 1C. As can be seen, the C-terminal substrate binding lobe, roughly defined by subdomains VI-XII, shows a greater frequency of disease SNPs compared with common SNPs. (Table 1 and Fig. 1C). Pairwise correlation analysis (r = −0.1551, P = 0.0264) and a simulation study (as described in Methods) revealed that specific positions within the catalytic core, especially within the C-terminal lobe, are enriched in disease mutations (Fig. 1B and Table S2). A detailed description of all of the disease SNPs and their structural location is given in the SI Materials and Methods and Figs. S1–S6. In the following sections, we focus on the sites that harbor four or more disease SNPs, both in the N-lobe and the C-lobe.
N-Lobe
Subdomain I.
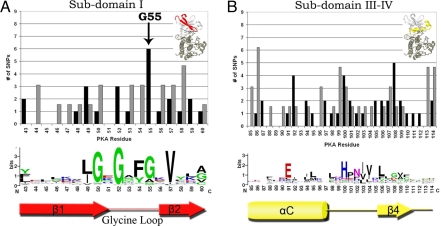
The most frequently mutated residue in subdomain I corresponds to a conserved glycine (G55) within the glycine rich G (50)XG(52)XXG (55) loop (G-loop) (Fig. 2A). The G-loop is one of the most flexible elements of the catalytic core and plays a key role in phosphoryl transfer. Specifically, G50 and G52 within the G-loop participate in the phosphoryl transfer reaction (16), and G55 plays a role in regulation, because it contributes to the conformational flexibility of the G-loop (17). It is notable that disease SNPs are enriched at sites involved in regulation rather than catalysis. In fact, mutation of G55 shows multiple effects on kinase activity. Replacement of G55 with valine or arginine decreases activity in INSR (G1035) (Table S3) (18), whereas substitutions of G55 by alanine or serine increase activity in BRAF (19) or leave activity unaffected in PKA (20).
Fig. 2.
Distribution of disease and common SNPs in N-lobe subdomains. The distribution of disease and common SNPs and the degree of conservation per residue in subdomains I (A) and III–IV (B). Black bars, disease SNPs; gray bars, common SNPs. The character height is proportional to the degree of conservation. The number of common SNPs is adjusted for the difference in total common and disease SNPs occurring throughout the catalytic core. Arrow denotes disease hotspot G55.
Subdomain III–IV.
Subdomain III–IV contains three residues frequently harboring disease SNPs (Fig. 2B). These correspond to K92 in the αC-helix, H100 (F in PKA) in the αC-β4 loop and F108 in the β4 strand. K92 is located in the flexible αC-helix, which serves as a docking site for regulatory proteins. In Cdk2, for instance, the K92 equivalent (I52) directly interacts with cyclin A, which is a key regulator of Cdk2 activity (20). Likewise, in AGC kinases, K92 positions the C-terminal tail, which serves as a cis-regulatory element (21). Moreover, K92 is strategically located relative to the kinase conserved E91, which positions the ATP by forming a salt bridge interaction with K72. Thus, mutation of K92 is likely to alter regulation either by decreasing catalytic activity as seen in INSR (A–D) (22), RSK2 (R–P) (23), and CYGD (F–S) (24), or constitutively activating the kinase as seen in KIT (K-E) (25).
H100 (F in PKA) is located in the αC-β4 loop, which anchors the flexible C-helix. H100 is part of the HxN motif, which is conserved in eukaryotic protein kinases (ePKs), but absent in distantly related eukaryotic-like kinases (ELKs) (26). This loop is the only part of the N-lobe that is firmly anchored to the C-lobe and serves as a hinge point for C-helix movement (26). More recent studies have suggested a role for this motif in kinase regulation (27), because variations within this loop appear to favor alternative modes of C-helix positioning (21, 28). Mutations at this site produce severe (29, 30) and/or dominant-negative effects (31).
F108 is located in the β4 strand, which forms a docking site for the C-tail in AGC kinases. F108 is specifically conserved in AGC kinases, but the precise role of this residue in AGC kinase functions is unclear.
C-Lobe.
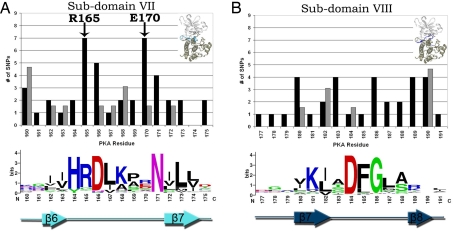
Subdomain VII.
Subdomain VII (Fig. 3A) contains key conserved residues that participate in diverse functions such as phosphoryl transfer, substrate binding, and regulation. Positions that harbor the most SNPs in this subdomain include the kinase conserved aspartate (D166) and asparagine (N171), involved in catalysis, the tyrosine kinase specific arginine R170 (E in PKA) implicated in substrate binding (32), and the regulatory arginine (R165) that coordinates with the phosphorylated residue in the activation loop. Notably, R165 and R170 are more frequently mutated in disease states compared with D166 and N171 (Tables S2 and S3). This, again, suggests that regulatory functions are more frequently altered in diseases states compared with catalytic functions, leading to variations in disease severity. For instance, in INSR, mutation of R1158 to tryptophan results in Rabson–Mendenhall syndrome (33), whereas mutation of the same arginine to glutamine results in Insulin resistance (34). Similarly, in ZAP70 mutation of R465 to histidine results in a selective T cell defect (35), whereas mutation of the same arginine to cysteine results in T-B- SCID (36). However, D166 mutations are characterized by a severe phenotype and lack of autophosphorylation activity (37, 38), and substitutions of N171 by lysine results in severe diseases such as Robinow syndrome or Coffin-Lowry syndrome (23, 39).
Fig. 3.
Distribution of disease and common SNPs in subdomains VII and VIII. The distribution of disease and common SNPs and the degree of conservation per residue in subdomains VII (A) and VIII (B). Black bars, disease SNPs; gray bars, common SNPs. The character height is proportional to the degree of conservation. The number of common SNPs is adjusted for the difference in total common and disease SNPs occurring throughout the catalytic core. Arrow denotes disease hotspots R165 and E170.
Subdomain VIII.
Subdomain VIII (Fig. 3B) also displays a similar trend where sites not directly involved in ATP binding or catalysis are more frequently altered in disease compared with the catalytic residues. Within the DFG motif, the DFG-aspartate, which chelates the magnesium ion, harbors only one disease SNP [D194N in LKB1 causing Peutz–Jeghers (40)], whereas the DFG-glycine, which contributes to the conformational flexibility of the DFG motif and the adjoining activation loop, is altered in four different kinases. Likewise, T183, which contributes to the conformational flexibility of the DFG motif by undergoing backbone torsion angle changes (26), and K189, which contacts the primary phosphorylation site in the activation loop (41), are also frequently altered in disease states. Movements of these residues, and the DFG+1 and DFG+2 residues (residues with no common polymorphisms), are required for adoption of the active conformation by rearranging disease-associated residues K189 and R165, building up the hydrophobic “spine,” and flipping the C-helix to secure the K72-E81 salt-bridge (42).
Subdomains IX–XII.
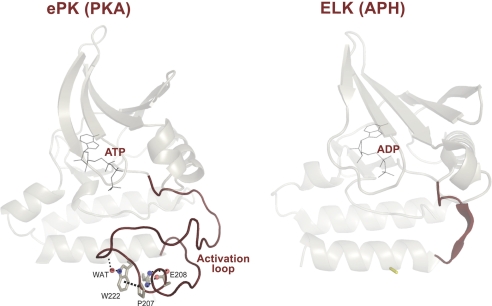
Subdomains IX–XII (Fig. S6) constitute the substrate binding region of the catalytic core defined by alpha helices F, G., H, and I. Although our knowledge of these subdomains is limited compared with subdomains in the N-terminal lobe, some studies have shown a role for the C-terminal subdomains in protein substrate interactions (43), tethering of substrates (44), and in allostery (45). The emerging theme from these studies is that tethering of substrates and regulatory proteins to distal sites in the C-lobe may help optimize catalysis at the active site. A recent comparative analysis of eukaryotic protein kinases (ePKs) and distantly related eukaryotic-like kinases (ELKs) demonstrated that key differences between ePKs and ELKs lie in the C-lobe of the catalytic core (Fig. 4) (46). In particular, the P+1 pocket in the activation segment and all of the key residues that anchor this pocket to the C-lobe were shown to be absent in ELKs. Because the P+1 pocket structurally links the subdomains in the C-lobe with the ATP and substrate binding regions in the N-lobe, the ePK specific network was suggested to play a role in ePK allostery (26). Surprisingly, the P+1 pocket and the residues that anchor this pocket are some of the most enriched in disease-associated mutations.
Fig. 4.
Conserved core shared between EPKs and ELK and structural elements unique to EPK. Structural comparison of PKA and distantly related aminoglycoside kinase (APH). A conserved core shared between PKA and APH is shown (in transparent mode). The exaggerated activation segment, which is one of the distinguishing features of EPKs is shown in red. The C-terminal substrate binding regions of PKA and APH are omitted for clarity. These C-terminal regions are very different and likely contribute to substrate specificity. The buried side-chain network that anchors the flexible activation segment is also shown. This network appears to have coevolved with the activation segment as ELKs that lack an exaggerated activation segment also lack the side-chain network.
Subdomain IX.
The P+1 motif, located in subdomain IX (Fig. S6A), is roughly defined by residues G200–E208 in the activation segment and contains the conserved APE motif. This segment is critical not only for substrate recognition but also as the hydrophobic glue that holds the subdomains of the C-lobe together. Throughout the catalytic core, the highest concentration of disease-associated residues occurs within the P+1 pocket. Residues G200 and T201, directly at the site of catalysis, are not significantly disease associated, whereas residues 203–208 are. Of these residues, E203 and L205 directly interact with substrates, whereas Y204 and the APE motif (A206, P207, and E208) do not. Y204 hydrogen bonds to E230 in the F-helix, which directly interacts with the peptide substrate in PKA, however, mutagenesis has revealed that the primary role of Y204 is to provide a hydrophobic surface to mediate allosteric regulation across the C-lobe (47). The APE motif, likewise, may be involved in this allosteric regulation, because it is anchored to the F, G, and I helices (discussed below), thereby providing direct communication between the activation segment and C-terminal subdomains. APE-glutamate, E208, is the only conserved electrostatic interaction that serves to stabilize cross communication across the C-Lobe and is a major hotspot for disease mutations (Table S3).
Subdomain X.
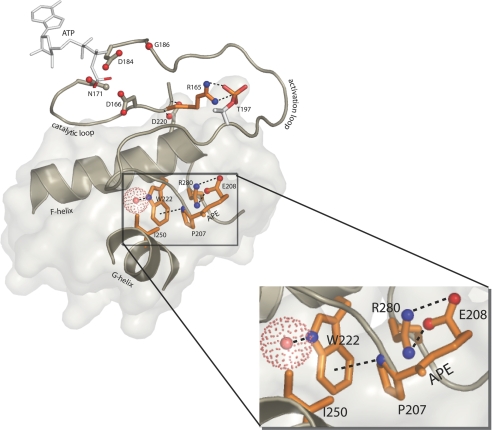
Subdomain X (Fig. S6C) contains the hydrophobic F Helix. This completely buried helix, an unusual element in soluble globular proteins, constitutes the “core” of the C-lobe to which every other C-lobe subdomain is anchored. Many hydrophobic residues in this helix are disease-associated, the most prominent of which is W222 (Table S3). W222 mediates a CH-pi interaction with the proline of the APE motif, positions the backbone of the APE motif via a conserved water molecule (46) (Fig. 5).
Fig. 5.
SNPs and allostery. The ePK conserved allosteric network of the C-terminal lobe. Red spheres, oxygen; blue spheres, nitrogen; dashed lines, hydrogen bonds. Zoom box shows the ePK conserved side-chain network.
Subdomains XI-XII.
Subdomains XI-XII, defined by helices G, H, and I (Figs. S4 and S6), are sparsely populated by disease-causing SNPs. The exception is R280, which is located between the H and I helices, and mutated in seven distinct kinases (Table S3). R280 forms a salt bridge interaction with the glutamate of the APE motif packs up against the W222 in the F-helix (Fig. 5). Mutation of this arginine to a lysine reduces catalytic activity in PKA but does not alter the overall structure or fold of the kinases (J. Yang and S.S.T., unpublished results).
Discussion
Our results indicate that perturbed kinase residues involved in functional regulation, allosteric networks, and substrate binding, especially residues indirectly involved in protein–protein interactions and allostery, are extremely important contributors to human disease caused by germ-line SNPs associated mostly with developmental and metabolic disorders (Table S4). In contrast, SNPs resulting in disease do not occur frequently at residues directly involved in catalysis, probably because perturbations at these highly conserved sites a complete loss of function and are only likely to occur in proteins whose functions are not essential for survival. It should be noted that the majority of diseases represented in the protein kinase family are developmental defects caused by loss of function mutations. Thus, preponderance of disease SNPs observed in kinases occur at sites where partial activity is conserved, and viability is retained, albeit often with severe biological deficits. This also suggests that the disease SNPs occurring in heterozygous states act in a haploinsufficient or dominantly negative manner. It is possible that the greater frequency of disease SNPs at regulatory or substrate binding sites, rather than catalytic sites, may be a general property of disease SNPs in other catalytic enzymes-the largest class of disease-causing proteins (48).
Our analyses reveal that hotspots for disease SNPs occur at sites conserved across species in eukaryotic protein kinases (ePKs) and not in the prokaryotic eukaryotic-like kinases (ELKs) (46) and are likely to be involved in functions specific to ePKs. Of 10 key residues conserved across ePKs and ELKs-G52, K72, E91, P104, H158, H164, D166, N171, D184, and D220 (46), only D166 is among the top 10 disease-associated residues. These results are consistent with the recent results of a survey of functional genomic elements by the ENCODE Project Consortium (49). The ENCODE researchers identified a number of regions of the genome that exhibited clear biological activities but were not conserved across species, suggesting a role for lineage-specific variations in mediating particular biological functions.
It is these lineage-specific functions, built on top of the more ancient catalytic machinery, that appear to be the major target of disease SNPs. For example, the highly disease-associated residues of the N-lobe: The third glycine of the G-loop (G55), the histidine of the HxN motif (H100), and the putative regulatory molecule docking sites K92 and F108, which cap the αC-β4 region, have been shown (G55, K92, and H100) or are likely (F108) to be key players in movements of the C-helix from the inactive to active conformation in ePKs (Fig. S5). In contrast, the C-helix is held in a constitutively active conformation in ELKs. Anchoring of the C-helix is a key regulatory element in ePKs.
C-helix movements, an N-lobe ePK specific function, are also influenced by regulatory events in the C-lobe, such as movement of subdomain VIII. However, the majority of disease hot spot residues are involved in the side-chain network formed by the APE motif, W222 and R280 (Fig. 5), recently shown to be a unique feature of ePKs (46). Distantly related ELKs in prokaryotes that phosphorylate small metabolites lack these residues (26), suggesting a role for the ePK-specific network in substrate binding function and allosteric regulation. Consistent with this notion, mutation of the APE glutamate to lysine in ILK may reduces substrate affinity (50), or alternatively, may reduce affinity for the associated kinase responsible for substrate phosphorylation (51). Likewise, mutation of the arginine of subdomain XII in yeast PKA was shown to affect binding and release of protein substrates (52). It is interesting that, although they are not exposed to solvent, these residues are indirectly contributing to substrate binding. Further characterization of these residues is required to precisely understand the role of these residues in protein kinase structure, function, and disease.
Ultimately, our results could not have been anticipated without an in-depth study of the unique evolutionary and functional features of kinases and hence extends the findings of research that considers general or ubiquitous sequence-based features of nsSNPs (10, 11). In this light, we can speculate about kinase SNPs that may cause disease by extrapolating our results and hypothesize that SNPs within the coding regions of kinase genes could influence common disease if they occur at positions that mildly affect substrate binding or allosteric regulation—especially if they appear to be lineage-specific residues—even though their ultimate functional affects may not be immediately obvious without structural or functional characterization. A major challenge for the future will be to delineate the role of SNPs within individual kinase families using computational and experimental methods.
Methods
Disease-causing and common SNPs were obtained and mapped to kinase sequences as described in ref. 12 (see SI Materials and Methods). A nonredundant set of SNPs was generated from all ePK genes carrying a disease-causing or common SNP, so that no site within a particular kinase was counted more than once. In total, 428 disease-causing SNPs and 330 common SNPs were compiled for the analyses. The majority of disease-causing SNPs are derived from targeted sequencing of kinase genes of interest for germ-line mutations in patient families with a specific disease, whereas the majority of common SNPs are derived from large-scale sequencing efforts. Kinase sequences were aligned to characteristic catalytic site motifs. These alignments, using all human ePK sequences harboring common or disease-causing mutations, were used to generate all logo figures, using WebLogo (53). Regions are denoted based on the definitions provided by Hanks and Hunter (13), where a denotes the intervening region between subdomains. Note that subdomain X is split in two halves, X(i) and X(ii). For a detailed description of the characteristics of the subdomains and their resident conserved amino acids, see Hanks and Hunter (13).
The expected probability [E(p)] of a SNP occurring in a region was calculated separately for common and disease SNPs as follows: The average length of each region was calculated as the weighted average of the region length in each kinase considered, where weights correspond to the total number of SNPs occurring within each kinase. This weighting helps avoid biases that might arise as a result of some kinases simply harboring more SNPs than others. The probability of a SNP occurring within a particular region purely by chance was computed as its weighted average length over the sum of every region's weighted average length.
The probability (P value) of the observed total number (x) of SNPs occurring within each region, where n is the total number of SNPs considered, was calculated using the general binomial distribution as follows: If x/n < E(p):
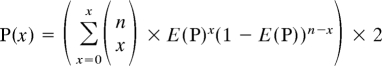 |
If x/n > E(p):
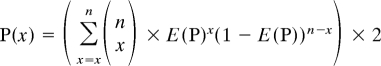 |
Comparisons of the average length per region in the common and disease SNPs sets, the comparison of the number of SNPs per region, and the number occurring within subdomains vs. intervening regions were calculated by using the normal distribution approximation to the binomial distribution.
Multiple alignments were generated by using a motif model. Sites with multiple disease SNPs were considered for further structural analysis. To estimate whether disease SNPs are position-specific or distributed randomly throughout the catalytic domain, in addition to a pairwise correlation, we ran 10,000 Monte Carlo simulations involving random assignment of disease SNPs. The SNP distribution resulting from this simulation study compared with the observed distribution was that zero SNPs occurred at an average of 19.52 ± 0.03 positions in the simulation vs. 46 observed positions; one SNP at 67.58 ± 0.06 positions vs. 65 observed positions; two SNPs at 76.95 ± 0.07 positions vs. 47 observed positions; three SNPs at 35.04 ± 0.04 positions vs. 18 observed positions; four SNPs at 7.20 ± 0.03 positions vs. 21 observed positions, five SNPs at 0.69 ± 0.01 positions vs. 3 observed positions; six SNPs at 0.03 ± 0.002 positions vs. 3 observed positions; seven SNPs at 0.0002 ± 0.0001 positions vs. 3 observed positions; and eight SNPs at 0.0 ± 0.0 positions vs. 1 observed position. Thus, the observed distribution is enriched for position specific mutations at positions where four or more mutations are observed.
Supplementary Material
Acknowledgments.
We thank Dr. Gerard Manning for invaluable discussions regarding the manuscript. This work was supported in part by National Heart Lung and Blood Institute Family Blood Pressure Program Grant U01 HL064777-06, National Institute on Aging Longevity Consortium Grant U19 AG023122-01, National Institute of Mental Health Consortium on the Genetics of Schizophrenia Grant 5 R01 HLMH065571-02, National Institute of Health Grants R01 HL074730-02 and R01 HL070137-01, and Scripps Genomic Medicine (to N.J.S.); a University of California at San Diego Genetics Training Grant for the Biomedical Sciences (to A.T.); and National Institute of Health Grant GM19301 (to N.K. and S.S.T.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802403105/DCSupplemental.
References
- 1.Knighton DR, et al. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent kinase. Science. 1991;253:407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T. The croonian lecture 1997 The phosphorylation of proteins on tyrosine: Its role in cell growth and disease. Philos Trans R Soc Lond B. 1998;353:583–605. doi: 10.1098/rstb.1998.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 4.Huang H. Evolutionary conservation and selection of human disease gene orthologs in the rat and mouse genomes. Genome Biol. 2004;5:R47. doi: 10.1186/gb-2004-5-7-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ortutay C, Valiaho J, Stenberg K, Vihinen M. KinMutBase: A registry of disease-causing mutations in protein kinase domains. Hum Mutat. 2005;25:435–442. doi: 10.1002/humu.20166. [DOI] [PubMed] [Google Scholar]
- 6.Stenson PD, Ball EV, Shiel AD, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 7.Hamosh A, Scott AF, Amberger JS, Carol AB, McKusick VA. Online Medelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lohmueller KE, et al. Proportionally more deleterious genetic variation in European than in African populations. Nature. 2008;451:994–997. doi: 10.1038/nature06611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newton-Cheh C, Hirschorn JN. Genetic association studies of complex traits: design and analysis issues. Mutat Res. 2005;573:54–69. doi: 10.1016/j.mrfmmm.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 10.Ng PC, Henikoff S. Predicting the effects of amino Acid substitutions on protein function. Ann Rev Gen Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- 11.Torkamani A, Schork AT. Accurate prediction of deleterious protein kinase polymorphisms. Bioinformatics. 2007;23:2918–2925. doi: 10.1093/bioinformatics/btm437. [DOI] [PubMed] [Google Scholar]
- 12.Torkamani A, Schork NJ. Distribution analysis of nonsynonymous polymorphisms within the human kinase gene family. Genomics. 2007;90:49–58. doi: 10.1016/j.ygeno.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Hanks SK, Hunter T. Protein kinases 6 The eukaryotic protein-kinase superfamily- kinase (catalytic) domain-structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 14.Torkamani A, Schork NJ. Prediction of cancer driver mutations in protein kinases. Cancer Res. 2008;68:1675–1682. doi: 10.1158/0008-5472.CAN-07-5283. [DOI] [PubMed] [Google Scholar]
- 15.Niedner RH, et al. Protein kinase resource: An integrated environment for phosphorylation research. Proteins. 2006;63:78–86. doi: 10.1002/prot.20825. [DOI] [PubMed] [Google Scholar]
- 16.Hemmer W, McGlone M, Tsigelny I, Taylor SS. Role of the glycine triad in the atp-binding site of cAMP-dependent protein kinase. J Biol Chem. 1997;27:16946–16954. doi: 10.1074/jbc.272.27.16946. [DOI] [PubMed] [Google Scholar]
- 17.Grant BD, Hemmer W, Tsigelny I, Adams JA, Taylor SS. Kinetic analyses of mutations in the glycine-rich loop of cAMP-dependent protein kinase. Biochemistry. 1998;37:7708–7715. doi: 10.1021/bi972987w. [DOI] [PubMed] [Google Scholar]
- 18.Odawara M, et al. Human diabetes associated with a mutation in the tyrosine kinase domain of the insulin receptor. Science. 1989;245:66–68. doi: 10.1126/science.2544998. [DOI] [PubMed] [Google Scholar]
- 19.Ikenoue T, et al. Different effects of point mutations within the B-Raf glycine-rich loop in colorectal tumors on mitogen-activated protein/extracellular signal-regulated kinase kinase/extracellular signal-regulated kinase and nuclear factor κB pathway and cellular transformation. Cancer Res. 2004;64:3428–3435. doi: 10.1158/0008-5472.CAN-03-3591. [DOI] [PubMed] [Google Scholar]
- 20.Jeffery D, et al. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 21.Kannan N, Haste N, Taylor SS, Neuwald AF. The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proc Nat Acad Sci. 2007;103:1272–1277. doi: 10.1073/pnas.0610251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haruta T, et al. Ala1048 → Asp mutation in the kinase domain of insulin receptor causes defective kinase activity and insulin resistance. Diabetes. 1993;42:1837–1844. doi: 10.2337/diab.42.12.1837. [DOI] [PubMed] [Google Scholar]
- 23.Delaunoy J, et al. Mutations in the X-linked RSK2 gene (RPS6KA3) in patients with Coffin–Lowry syndrome. Hum Mutat. 2001;17:103–116. doi: 10.1002/1098-1004(200102)17:2<103::AID-HUMU2>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 24.Perrault I, et al. Retinal-specific guanylate cyclase gene mutations in Leber's congenital amaurosis. Nat Genet. 1996;14:461–464. doi: 10.1038/ng1296-461. [DOI] [PubMed] [Google Scholar]
- 25.Isozaki K, et al. Germline-activating mutation in the kinase domain of KIT gene in familial gastrointestinal stromal tumors. Am J Pathol. 2000;157:1581–1585. doi: 10.1016/S0002-9440(10)64795-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kannan N, Neuwald AF. Did protein kinase regulatory mechanisms evolve through elaboration of a simple structural component? J Mol Biol. 2005;351:956–972. doi: 10.1016/j.jmb.2005.06.057. [DOI] [PubMed] [Google Scholar]
- 27.Kannan N, Neuwald AF, Taylor SS. Analogous regulatory sites within the alphaC-beta4 loop regions of ZAP-70 tyrosine kinase and AGC kinases. Biochim Biophys Acta. 2008;1784:27–32. doi: 10.1016/j.bbapap.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deindl S, et al. Structural basis for the inhibition of tyrosine kinase activity of ZAP-70. Cell. 2007;129:735–746. doi: 10.1016/j.cell.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 29.Ezoe K, et al. Novel mutations and deletions of the KIT (steel factor receptor) gene in human piebaldism. Am J Hum Genet. 1995;56:58–66. [PMC free article] [PubMed] [Google Scholar]
- 30.Hatano Y, et al. Novel PINK1 mutations in early-onset parkinsonism. Ann Neurol. 2004;56:424–427. doi: 10.1002/ana.20251. [DOI] [PubMed] [Google Scholar]
- 31.Murakami T, et al. Analysis of KIT, SCF, and initial screening of SLUG in patients with piebaldism. J Invest Dermatol. 2005;124:670–672. doi: 10.1111/j.0022-202X.2005.23637.x. [DOI] [PubMed] [Google Scholar]
- 32.Hubbard SR. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 1997;16:5572–5581. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Longo N, et al. Genotype-phenotype correlation in inherited severe insulin resistance. Hum Mol Genet. 2002;11:1465–1475. doi: 10.1093/hmg/11.12.1465. [DOI] [PubMed] [Google Scholar]
- 34.Kishimoto M, et al. Substitution of glutamine for arginine 1131 A newly identified mutation in the catalytic loop of the tyrosine kinase domain of the human insulin receptor. J Biol Chem. 1994;269:11349–11355. [PubMed] [Google Scholar]
- 35.Toyabe S-I, Watanabe A, Harada W, Karasawa T, Uchiyama M. Specific immunoglobulin E responses in ZAP-70-deficient patients are mediated by Syk-dependent T-cell receptor signalling. Immunology. 2001;103:164–171. doi: 10.1046/j.1365-2567.2001.01246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elder ME, et al. Distinct T cell developmental consequences in humans and mice expressing identical mutations in the DLAARN motif of ZAP-70. J Immunol. 2001;166:656–661. doi: 10.4049/jimmunol.166.1.656. [DOI] [PubMed] [Google Scholar]
- 37.Mehenni H, et al. Loss of LKB1 kinase activity in Peutz–Jeghers syndrome, and evidence for allelic and locus heterogeneity. Am J Hum Genet. 1998;63:1641–1650. doi: 10.1086/302159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hashimoto S, et al. Identification of Bruton's tyrosine kinase (Btk) gene mutations and characterization of the derived proteins in 35 X-linked agammaglobulinemia families: A nationwide study of Btk deficiency in Japan. Blood. 1996;88:561–573. [PubMed] [Google Scholar]
- 39.Afzal AR, et al. Recessive Robinow syndrome, allelic to dominant brachydactyly type B, is caused by mutation of ROR2. Nat Genet. 2000;25:419–422. doi: 10.1038/78107. [DOI] [PubMed] [Google Scholar]
- 40.Westerman AM, et al. Novel mutations in the LKB1/STK11 gene in Dutch Peutz–Jeghers families. Hum Mut. 1999;13:476–481. doi: 10.1002/(SICI)1098-1004(1999)13:6<476::AID-HUMU7>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 41.Nolen B, Taylor SS, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 42.Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci USA. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dar AC, Dever TE, Sicheri F. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell. 2005;122:887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 44.Lee T, et al. Docking motif interactions in MAP kinases revealed by hydrogen exchange mass spectrometry. Mol Cell. 2004;14:43–55. doi: 10.1016/s1097-2765(04)00161-3. [DOI] [PubMed] [Google Scholar]
- 45.Hantschel O, et al. A myristoyl/phosphotyrosine switch regulates c-Abl. Cell. 2003;112:845–857. doi: 10.1016/s0092-8674(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 46.Kannan N, Taylor SS, Zhai Y, Venter JC, Manning G. Structural and Functional Diversity of the Microbial Kinome. PLoS Biol. 2007;5:e17. doi: 10.1371/journal.pbio.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J, et al. Allosteric network of cAMP-dependent protein kinase revealed by mutation of Tyr204 in the P+1 Loop. J Mol Bio. 2005;346:191–201. doi: 10.1016/j.jmb.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 48.Jimenez-Sanchez G, Childs B, Valle D. Human disease genes. Nature. 2001;409:853–855. doi: 10.1038/35057050. [DOI] [PubMed] [Google Scholar]
- 49.ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carnio LK. Toronto: University of Toronto; 2000. PhD Thesis. [Google Scholar]
- 51.Lynch DK, Ellis CA, Edwards PA, Hiles ID. Integrin-linked kinase regulates phosphorylation of serine 473 of protein kinase B by an indirect mechanism. Oncogene. 1999;18:8024–8032. doi: 10.1038/sj.onc.1203258. [DOI] [PubMed] [Google Scholar]
- 52.Deminoff SJ, Howard SC, Hester A, Warner S, Herman PK. Using substrate-binding variants of the cAMP-dependent protein kinase to identify novel targets and a kinase domain important for substrate interactions in Saccharomyces cerevisiae. Genetics. 2006;173:1909–1917. doi: 10.1534/genetics.106.059238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moreland JL, Gramada A, Buzko OV, Zhang Q, Bourne PE. The molecular biology toolkit (mbt): A modular platform for developing molecular visualization applications. BMC Bioinformatics. 2005;6:21. doi: 10.1186/1471-2105-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.