Abstract
The hemagglutinin-esterases (HEs) are a family of viral envelope glycoproteins that mediate reversible attachment to O-acetylated sialic acids by acting both as lectins and as receptor-destroying enzymes (RDEs). Related HEs occur in influenza C, toro-, and coronaviruses, apparently as a result of relatively recent lateral gene transfer events. Here, we report the crystal structure of a coronavirus (CoV) HE in complex with its receptor. We show that CoV HE arose from an influenza C-like HE fusion protein (HEF). In the process, HE was transformed from a trimer into a dimer, whereas remnants of the fusion domain were adapted to establish novel monomer–monomer contacts. Whereas the structural design of the RDE-acetylesterase domain remained unaltered, the HE receptor-binding domain underwent remodeling to such extent that the ligand is now bound in opposite orientation. This is surprising, because the architecture of the HEF site was preserved in influenza A HA over a much larger evolutionary distance, a switch in receptor specificity and extensive antigenic variation notwithstanding. Apparently, HA and HEF are under more stringent selective constraints than HE, limiting their exploration of alternative binding-site topologies. We attribute the plasticity of the CoV HE receptor-binding site to evolutionary flexibility conferred by functional redundancy between HE and its companion spike protein S. Our findings offer unique insights into the structural and functional consequences of independent protein evolution after interviral gene exchange and open potential avenues to broad-spectrum antiviral drug design.
Keywords: sialic acid, x-ray crystallography, glycoprotein, lectin, nidovirus
Many different respiratory and enteric viruses initiate infection by binding to sialic acids. These nine-carbon amino sugars are common terminal residues of glycoconjugates and occur in a wide variety, which arises from differential modification of the parental molecule, 5-N-acetyl neuraminic acid (Neu5Ac), and from differences in glycosidic linkage (1). Although viruses may attain host and cell specificity by being selective for particular sialic acid subtypes (1–4), even these receptors may still occur abundantly on off-target host cells (including the infected cell), on viral progeny, and on soluble and insoluble molecules in the extracellular environment. To prevent irreversible binding to “decoy receptors” and consequent loss of infectivity, orthomyxo-, paramyxo-, and coronaviruses encode virion-associated receptor-destroying enzymes (RDEs) (5–7). In turn, RDEs make prime targets for antiviral intervention, as exemplified by the influenza A virus neuraminidase (8–10).
The coronaviruses are a group of enveloped (+)strand RNA viruses that comprises several important pathogens of human clinical and veterinary importance, including severe acute respiratory syndrome (SARS)-coronavirus, human respiratory coronavirus (HCoV) OC43 and its proposed zoonotic ancestor (11), bovine coronavirus (BCoV). HCoV-OC43 and BCoV use 9-O-acetylated sialic acids as receptor and correspondingly possess sialate-9-O-acetylesterases as RDE (12, 13). Enzymatic activity is associated with the hemagglutinin-esterase (HE), a homodimeric class I membrane protein (for a review, see ref. 7). Although among coronaviruses, HE is exclusive to members of phylogenetic subgroup 2a (7), closely related proteins do occur in toroviruses (14–16) and in orthomyxoviruses of mammals (17) and fish (18). Best studied is the influenza C virus HE fusion protein (HEF), a homotrimeric multifunctional spike protein that mediates virion attachment, receptor destruction, and membrane fusion (19–22).
The presence of homologous HE genes in the genomes of viruses from three distinct taxa is generally ascribed to RNA recombination and would have required two lateral gene transfer events at the minimum (7, 14, 23). The source of the HE module and the direction of gene flow remain to be established, however (7). Moreover, the influenza, toro-, and coronavirus HEs followed independent evolutionary trajectories, the functional and structural consequences of which are poorly understood. Unlike HEF, CoV HEs lack membrane fusion activity and, in the virion, they are accessory to the spike protein S, a receptor-binding fusion protein. Furthermore, although CoV HEs do bind to sialic acid, their role in virion attachment is subject to debate (7). Reportedly, CoV HEs serve primarily as RDE (24), but this view is at variance with observations that esterase-deficient HE promotes viral spread in vivo (25). Intriguingly, there is no obvious sequence homology between the HEF receptor-binding site and corresponding regions in HE, raising the question of whether CoV HEs possess a separate receptor-binding site or mediate sialic acid-binding and catalysis through a single site in the esterase pocket (7).
To further our understanding of the function and evolution of coronavirus HEs, we expressed BCoV HE and solved its structure, free and in complex with its receptor.
Results and Discussion
Expression, Purification, and Biochemical Analysis of CoV HE.
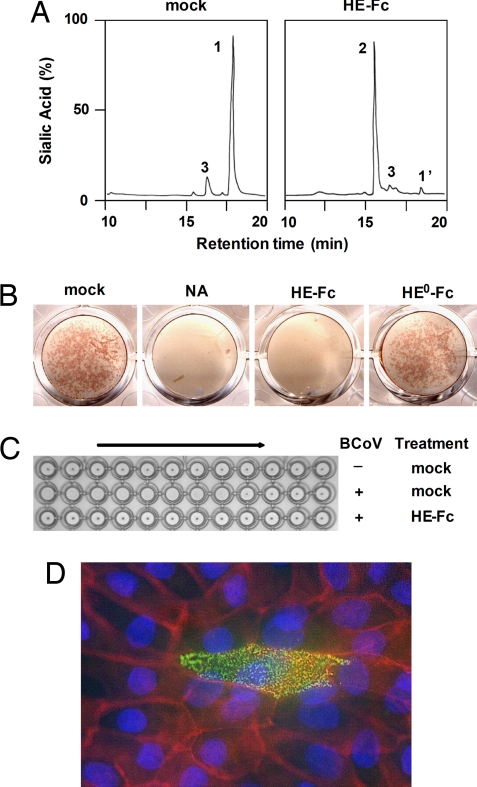
We produced the ectodomain of BCoV HE (residues 19–388) in enzymatically active (HE) and inactive form (HE0, containing an active site Ser40-to-Ala substitution) as Fc-fusion proteins by transiently transfecting N-acetylglucosaminyltransferase-deficient HEK293S cells with expression plasmids encoding the respective molecules. Soluble CoV HE was purified by protein A-affinity chromatography of the Fc chimeras followed by on-the-bead thrombin digestion. The proteins were tested in a series of assays to confirm that biological functions had been retained also as an indication for proper folding. Indeed, in cleavage assays with a synthetic di-O-acetylated Sia (5-N-acetyl-4,9-di-O-acetylneuraminic acid α-methylglycoside, αNeu4,5,9Ac32Me) as substrate, HE-Fc displayed sialate-9-O-acetylesterase activity (Fig. 1A). In addition, HE-Fc, but not its esterase-deficient derivative HE0-Fc, specifically destroyed BCoV receptors on rat erythrocytes and on Madin–Darby bovine kidney (MDBK) cells (Fig. 1 B and C). Moreover, HE-Fc and HE0-Fc retained lectin activity as demonstrated by hemagglutination assay (Figs. 1C and 4E) and by Sia-specific immunofluorescence staining of tissue culture cells (Fig. 1D).
Fig. 1.
BCoV HE-Fc chimeras retain esterase and lectin activity. (A) HE-Fc displays sialate-9-O-acetylesterase activity. Purified HE-Fc was assayed for substrate specificity as described (16) with the synthetic di-O-acetylated sialic acid analogue αNeu4,5,9Ac32Me as substrate. Graphs show total ion current gas chromatograms. Sialic acids were identified by mass spectrometry: αNeu4,5,9Ac32Me (peak 1), αNeu4,5Ac22Me (peak 2); peaks 1′ and 3 represent nonsialic acid compounds. (B) HE-Fc, but not esterase-deficient HE0-Fc, destroys BCoV receptors on the surface of tissue culture cells. MDBK cell monolayers were mock-treated, treated with neuraminidase (NA), or with the HE-Fc chimeras and subsequently infected with BCoV. Infected cells were visualized by an in situ acetylesterase staining. (C) HE-Fc destroys BCoV receptors on rat erythrocytes. Erythrocytes were mock-treated or treated with HE-Fc and subsequently subjected to hemagglutination assay with two-fold dilutions of BCoV (+) or with PBS (−). (D) HE0-Fc lectin activity allows immunohistochemical detection of 9-O-acetylated sialic acid on monolayers of polarized MDBK cells (green). Cell contacts were visualized by staining for PAN-Cadherin (red) and nuclei by staining with HOECHST 33258 (blue).
Fig. 4.
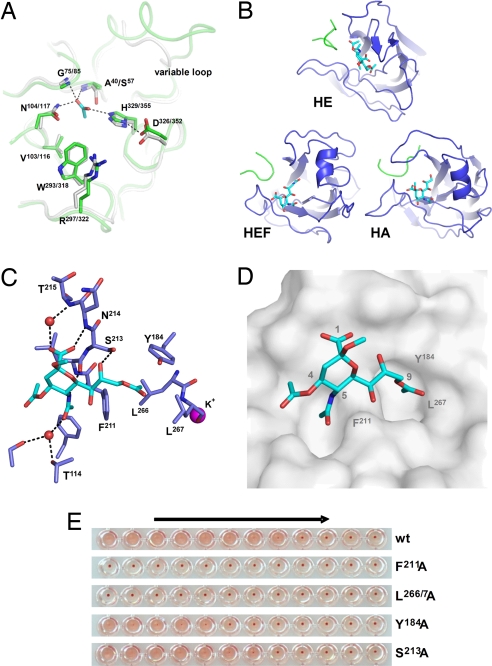
HE and HEF display conserved enzymatic but divergent receptor-binding sites. (A) Superposition of active site residues of HE (green) and HEF (gray). An acetate ion occupying the oxyanion hole in HE is shown in stick representation (carbon, cyan; oxygen, red). Catalytic triad and oxyanion hole hydrogen bonds are indicated by dashed black lines. Also shown is an adjacent loop that is highly variable also among CoVs. (B) Comparison of the receptor-binding regions of HE, HEF, and HA. Bound ligands, αNeu4,5,9Ac32Me in HE, αNeu5,9Ac22Me in HEF, and αNeu5Ac2Me in HA are shown in stick representation (carbon, cyan; nitrogen, blue; oxygen, red). Proteins are depicted in similar orientations obtained by superposition of their conserved central cores. (C) Ligand bound to the HE receptor-binding site in stick representation. Water molecules are shown as red spheres, a potassium ion as magenta sphere. Hydrogen bonds with receptor are shown as dashed black lines. (D) Surface representation of the HE receptor-binding site revealing two pockets accommodating the 9-O- and 5-N-acetyl groups of the receptor. (E) The effect of Ala substitutions on receptor binding. Relative binding affinity of wild-type HE and its derivatives was assessed by hemagglutination assay with rat erythrocytes and twofold serial dilutions of each of the HE-Fc chimeras (1,000 to 0.5 ng of per well, arrow).
Structure Determination and Overall Structure.
Crystals of free HE and of a complex of HE0 with αNeu4,5,9Ac32Me diffracted to 2.1 and 1.8 Å resolution, respectively. The structures were solved by molecular replacement by using influenza C HEF residues 35–410 as search model (for detailed crystallographic statistics, see Table 1).
Table 1.
Data collection and refinement statistics
| BCoV HE without ligand | BCoV HE S40A with ligand | |
|---|---|---|
| Spacegroup | P6522 | P6522 |
| Cell dimension, Å | 88.8, 88.8, 282.4 | 89.3, 89.3, 280.4 |
| Resolution, Å* | 2.1–30 (2.10–2.21) | 1.8–50 (1.80–1.90) |
| Completeness, % | 99.6 (99.9) | 97.2 (86.6) |
| Multiplicity | 8.4 (8.5) | 20.7 (19.0) |
| Rmerge % | 13.5 (80.4) | 9.2 (86.6) |
| I/σ | 12.8 (2.5) | 25.5 (3.5) |
| No. of reflections | 37,791 | 57,018 |
| Rfactor/Rfree, % | 18.0/21.2 | 17.0/18.8 |
| Average B-factors, Å2 | ||
| Protein | 35.1 | 34.4 |
| Ligand | 45.2 | |
| Water | 45.5 | 39.7 |
| rmsd bond lengths, Å | 0.017 | 0.012 |
| rmsd bond angles, ° | 1.6 | 1.3 |
| No. of protein atoms (monomer) | 2,860 | 2,860 |
| No. of N-glycosylation sites | 6 | 6 |
| No. of glycan units | 7 | 7 |
| No. of waters | 257 | 283 |
*Values between brackets refer to the highest-resolution shell of data.
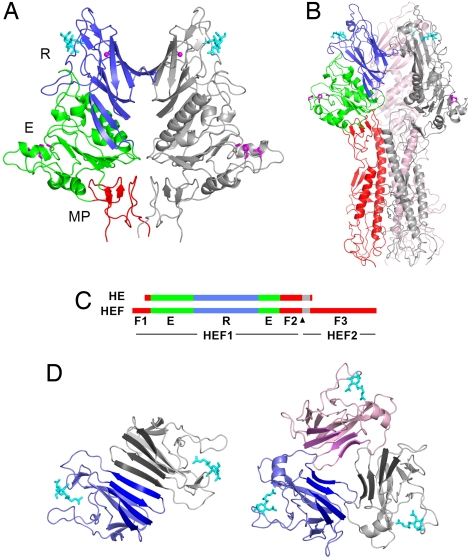
The data reveal that the CoV HE monomer is composed of three modules, each with an equivalent in influenza C HEF subunit 1 (HEF1): a receptor-binding domain (R), an acetylesterase domain (E), and a membrane-proximal domain (MP; Fig. 2 A–C). There is no equivalent of HEF subunit 2 (HEF2), which in HEF comprises the bulk of the fusion domain (F) (Fig. 2 B and C). Bound to the R domain is a well defined ligand molecule [supporting information (SI) Fig. S1] providing first and direct evidence for the presence of a genuine receptor-binding site. This observation unambiguously defines BCoV HE as a receptor-binding protein per se.
Fig. 2.
Structure of HE and comparison to HEF. (A) Ribbon representation of the BCoV HE dimer. One HE monomer is colored gray, the other by domain: lectin domain (R, blue) with bound αNeu4,5,9Ac32Me (cyan sticks) and potassium ion (magenta sphere); esterase domain (E, green) with Ser-His-Asp active site triad (magenta sticks); membrane-proximal domain (MP, red). (B) Structure of the influenza C HEF trimer. One monomer is colored by domain with domains R and E color-coded as in a; the fusion domain F is shown in red. Remaining monomers are shown in pink or gray. (C) Linear order of the sequence segments in HE and HEF color-coded by domains as in a and b. Gray segments indicate the transmembrane domain in HE and the fusion peptide in HEF. The arrowhead indicates the HEF cleavage site. (D) Top views showing the arrangement of the lectin domains in the BCoV HE homodimer (Left) and the influenza C HEF homotrimer (Right). The four-stranded β-sheets forming a continuous eight-stranded β-sheet in the HE dimer are emphasized by darker coloring.
In the virion, CoV HE forms dimers that are disulphide-linked through Cys385 near the C-terminal membrane anchor (7). This bond, although present in HE crystals (Fig. S2), was not visible in electron density maps because of disorder of residues 377–388. The biological dimer could, however, be identified uniquely from the proximity of C termini and size of interaction surface. It consists of identical molecules, related by twofold crystallographic symmetry, with an interface comprised of two major protein contact regions (CRs). CR1, associated with 952 Å2 buried solvent-accessible surface, involves a continuous eight-stranded β-sheet bridging the two R domains (Fig. 2D). CR2 buries 1,182 Å2 solvent-accessible surface and involves MP.
Origin and Evolution of CoV HE.
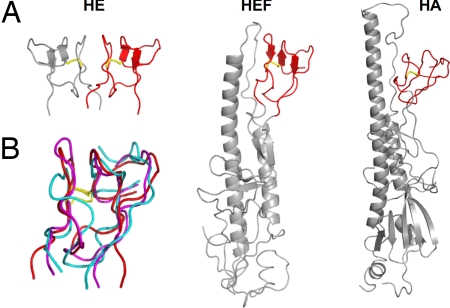
The structure of MP provides a decisive clue to the origin of CoV HE. As shown in Fig. 3, MP is strikingly similar to the F1/F2 globular region of HEF1 (22) and the F′ subdomain in the influenza A virus HA (26). In the native metastable forms of HEF and HA, these subdomains pack against a heptad-repeat loop that, as a prelude to membrane fusion, refolds into an α-helix at endosomal pH (26). HEF and HA are evolutionarily related but diverged long ago; their fusion domains are similar in structure, as are their R domains, but overall sequence identity has dwindled to only 12% (22). In contrast, HEF1 and CoV HE share 30% identity, indicative of a more recent split-off (7, 23). The presence in HE of an element that already must have been part of a fusion protein ancestral to HA and HEF (22) thus indicates that CoV HE originates from an HEF-like fusion protein and strongly argues against evolutionary models in which orthomyxo- and coronaviruses independently acquired an HE module solely comprised of R and E (7). In the simplest scenario (23), a group 2a CoV progenitor obtained through recombination the 5′-end of an HEF gene, including the coding sequence for the fusion peptide that came to serve as transmembrane domain (Fig. 2C). Without the HEF2 helical subdomain to direct trimerization, fortuitous interactions between transmembrane and/or F2 segments might have caused the HE protein to assemble into dimers instead; of note, the face of MP involved in dimer formation is equivalent to that of HEF segment F2 contacting the heptad repeat loop (Fig. 3). A four-stranded β-sheet in the R domain that remains unpaired in HEF and HA trimers (22) may have been remodeled subsequently to create a second, membrane-distal intermonomer contact to increase dimer stability (Fig. 2D).
Fig. 3.
MP is a remnant of a fusion domain. (A) Ribbon representations of MP in the HE dimer and of the fusion domains of the HEF and HA monomers. MP and corresponding subdomains in HEF and HA are shown in red; the remainder of the HEF and HA fusion domains are shown in gray. A conserved disulfide bond is shown in yellow stick representation. (B) Superposition of HE MP (red) and corresponding HEF (magenta) and HA subdomains (cyan). Sequence identity and rms deviations on Cα positions for HE-HEF and HE-HA are 33% and 1.9 Å, and 22% and 2.5 Å, respectively.
The Esterase Domain.
The E domain of CoV HE, harboring the RDE activity, has an SGNH-hydrolase fold (27) highly similar to that of HEF (1.4-Å rms deviation on Cα positions at 34% sequence identity). In fact, the active-site architecture of the HEF sialate-9-O-acetylesterase is fully conserved in BCoV HE with Ser40, His329, and Asp326 as catalytic triad and with the side chain of Asn104 and the NH groups of Ser40 and Gly75 forming an oxyanion hole (Fig. 4A). HE crystals soaked with 9-O-acetylated sialic acid did not contain this substrate in the enzymatic pocket. Instead, the oxyanion hole was occupied by an acetate ion, i.e., one of the catalytic products. The HEF and HE E domains do differ in size and structure of an extended surface loop (residues 47–54) that shields the catalytic pocket and that may prevent antibodies from accessing the functionally important esterase site (Fig. 4A). This loop is highly variable even among coronavirus HEs (7), tentatively identifying it as a site of antigenic variation.
A Receptor-Binding Site in CoV HE.
The R domain mediates receptor recognition (Fig. 1D). Although its core, an eight-stranded “Swiss roll,” is structurally well conserved and closely resembles that of HEF and HA (Fig. S3), its receptor-binding region is unique. This region is shaped by five variable loops, four grafted on the conserved core and one extending from the E domain. Differences between HE and HEF in loop size and conformation result in dramatically different binding sites that engage 9-O-acetylated sialic acid in nearly opposite orientations. Compared with the HEF–receptor complex, the receptor analogue bound to HE is rotated by 147° and shifted by 4 Å (Figs. 2D and 4B). Conversely, HEF and HA are similar with respect to receptor orientation and binding site topology (22) (Figs. 4B and Fig. S3). In HE, a β-hairpin, replacing an α-helix in HEF and HA interacts with the receptor through hydrogen bonds from main chain atoms of Leu212 and Asn214 to the 5-N-acetyl- and carboxylate groups, respectively, and from the side chain of Ser213 to O-8 (Fig. 4C). The α-2-O-methyl group of the receptor analogue points into the solvent, of relevance, because at this position, the natural ligand would be linked to the penultimate residue of the sugar chain. The 5-N-acetyl group fits into a pocket, its methyl interacting with a hydrophobic patch formed mainly by Phe211 (Fig. 4D). The 9-O-acetyl methyl of the receptor analogue docks into an adjacent hydrophobic pocket, formed by Tyr184, Phe211, Leu266, and Leu267 (Fig. 4 C and D), with the latter two residues in a loop stabilized by a potassium ion coordinated by Asp220, Ser221, Gln222, Ser263, Glu265, Leu267, and one water (Fig. S4). Given that the 9-O-acetyl moiety is a prerequisite for receptor binding and serves as a switch for virion attachment and release, this hydrophobic pocket must be key to ligand recognition. Indeed, Ala substitution of Phe211 or of Leu266 and Leu267 in tandem abrogated receptor binding, whereas substitution of Tyr184 decreased affinity significantly (Fig. 4E).
Plasticity of the CoV HE Receptor-Binding Site.
HEF also binds 9-O-acetylated sialic acid by contacting the 5-N-acetyl group through a hydrophobic patch and by burying the 9-O-acetyl in a nonpolar pocket (22). Therefore, despite the considerable differences in receptor-binding site architecture, HEF and HE use similar strategies to discriminate functional receptors from non-O-acetylated sialic acids. It is, however, surprising that the HE receptor-binding site has changed to this extent at all. Over a much larger evolutionary distance, the architecture of the HEF receptor-binding site has been preserved in HA, despite differences in receptor specificity (Neu5,9Ac2 vs. α2,3- or α2,6-linked Neu5Ac, respectively) and throughout antigenic variation (22, 26, 28) (Fig. 4B). Unlike HA and HEF, however, CoV HE acts in concert with another receptor-binding protein, S, a fusion protein essential for entry, and whereas HE-mediated virion attachment may be advantageous (25), it is not vital (16, 25, 29). We propose that redundancy in function between S and HE (24) afforded CoVs leeway to explore HE-binding site topologies and even to shift in coreceptor usage from 9-O- to 4-O-acetylated sialic acids in one branch of murine coronaviruses (16, 30–32).
The plasticity of the CoV HE receptor-binding site contrasts with the strict conservation of the E domain and the esterase catalytic site. From a pragmatic point of view, this may potentially allow the development of broad-spectrum antivirals targeting both orthomyxo- and coronavirus sialate-O-acetylesterases.
Materials and Methods
Construction of Expression Plasmids.
A synthetic codon-optimized sequence for the HE ectodomain of BCoV strain Mebus (amino acid residues 19–388) was cloned in a derivative of expression plasmid S1-Ig (33). The resulting construct, pCD5-BCoVHE-T-Fc, encodes a chimeric HE protein provided with an N-terminal CD5 signal peptide and at its C terminus, separated by a thrombin cleavage site, the Fc domain of human IgG1. To allow expression of an enzymatically inactive HE-Fc chimera (HE0-Fc), we generated a derivative of pCD5-BCoVHE-T-Fc in which the codon for the esterase catalytic residue Ser40 was substituted by Ala by site directed mutagenesis using the QuikChange XL II kit (Stratagene). Ala substitutions of key residues in the HE receptor-binding site were made likewise.
Protein Expression and Purification.
HEK293S cells lacking N-acetylglucosaminyl-transferase I (GnTI) activity (34) were transiently transfected with the expression plasmids using polyethyleneimine. At 18 h after transfection, the transfection mixture was replaced by 293 SFM II expression medium (Invitrogen), supplemented with sodium bicarbonate (3.7 g/liter), glucose (2.0 g/liter), Primatone RL-UF (3.0 g/liter), penicillin (100 units/ml), streptomycin (100 μg/ml), glutaMAX (Gibco), and 1.5% DMSO. Tissue culture supernatants were harvested 5–6 days after transfection, and HE-Fc and HE0-Fc were purified by protein A-affinity chromatography. For crystallography, the HE and HE0 ectodomains were cleaved off by on-the-bead thrombin digestion, concentrated to 10–15 mg/ml, and dialyzed against crystallization buffer (Tris·HCl, pH 8.2; 50 mM NaCl).
Biological and Biochemical Characterization of HE-Fc Chimeras.
Molecular weights of the proteins were determined by SDS/PAGE (under reducing and nonreducing conditions) and by gel filtration chromatography. O-acetylesterase activity was routinely measured by pNPA assay (19). Enzymatic de-O-acetylation of αNeu4,5,9Ac32Me was analyzed by GC-MS (16, 35). To assess esterase activity of the expression products toward cell-bound 9-O-acetylated sialic acids, MDBK cell monolayers in 16-mm diameter wells were treated with 20 μg/ml HE-Fc or HE0-Fc in PBS for 2 h at 37°C and subsequently infected with BCoV strain Mebus. Monolayers, mock-treated or treated with Vibrio cholerea neuraminidase (NA; 0.6 units/ml), were included as controls. Infected cells were visualized by staining using an in situ acetylesterase assay (36). Receptor-destroying activity toward 9-O-acetylated sialic acids on rat erythrocytes was assessed by incubating the cells with 20 μg/ml HE-Fc in PBS for 2.5 h at 37°C, before hemagglutination assay with twofold serial dilutions of BCoV. In situ detection of 9-O-acetylated sialic acids in cultured cells was demonstrated by performing a standard indirect immunofluorescence assay. Briefly, monolayers of polarized MDBK cells, grown on glass coverslips, were fixed with 2% paraformaldehyde and successively incubated with ≈25 μg/ml HE0-Fc in PBS, 0.05% Tween20 and with FITC-conjugated anti-human IgG1 (Nordic, 1:100) as secondary antibody. The samples were mounted on glass slides in FluorSave (Calbiochem) and examined by confocal fluorescence microscopy (Leica TCS SP2).
Crystallization.
Crystallization conditions were screened by the sitting-drop vapor diffusion method using a Honeybee 961 (Genomic Solutions). Drops were set up with 0.1 μl of HE protein solution in 10 mM Tris·HCl, pH 8.0, and 0 l reservoir solution. Diffracting crystals were obtained from JCSG solution D12 [0.04 M KH2PO4, 16% (wt/vol) PEG 8000, 20% (wt/vol) glycerol] (JCSG Technologies) and PEGIon-lite solution 89 [0.1 M KH2PO4, 10% (wt/vol) PEG 3350] (Hampton Research) at 18°C. Crystals for diffraction experiments were grown with the hanging-drop vapor diffusion method set up by hand with 0.8 μl of reservoir solution and 0.8 μl of protein solution at 18°C. Under both crystallization conditions, crystals of wild-type HE and HE0 grew within 2 weeks with the shape of hexagonal bipyramids to a size of ≈0.25 × 0.16 × 0.16 mm. Their space group is P6522. Crystals of wild-type HE grown at 0.1 M KH2PO4, 10% (wt/vol) PEG 3350 were flash-frozen in liquid nitrogen using reservoir solution with 22.5% (wt/vol) glycerol as the cryoprotectant.
Data Collection and Structure Solution.
To determine the HE structure in complex with its receptor analog Neu4,5,9Ac32Me (16), HE0 was used to prevent substrate degradation during soaking. Crystals obtained from 0.1 M KH2PO4, 10% (wt/vol) PEG 3350 cracked during ligand soaking and the flash-freezing procedure. Therefore, we used for substrate-soaking crystals of the mutant grown from 0.04 M KH2PO4, 16% (wt/vol) PEG 8000, 20% (wt/vol) glycerol, which did not require addition of cryoprotectant. Soaking was performed by adding 2 μl of reservoir solution with 10 mM Neu4,5,9Ac32Me directly into the margin of the drop with crystals, resulting in a final substrate concentration of ≈7 mM. Crystals were flash-frozen after 5–10 min.
Diffraction data of native and ligand-soaked crystals of BCoV HE were collected at European Synchrotron Radiation Facility station ID14-3 and ID 23-1, respectively. Diffraction data were processed by using XDS (37) and scaled using SCALA from the CCP4 suite (38). Molecular replacement was performed by using PHASER with Influenza C HEF residues 35–410 as template [Protein Data Bank (PDB) ID code 1FLC]. Density modification was carried out by using RESOLVE (PHENIX package) (39) and CNS (40). Approximately 280 residues were built automatically by ARP/WARP (41); the remaining residues were built manually with Coot (42). Refinement was carried out by using REFMAC (43); water molecules were added by using ARP/WARP. Molecular graphics were generated with PYMOL (http://pymol.sourceforge.net).
The structure of HE0 in complex with Neu4,5,9Ac32Me was solved by molecular replacement by using the wild-type HE structure as template. The structure was refined as described for wild-type HE. The library file for refinement of the ligand in REFMAC was made by adding chemical and geometric descriptions of the 4- and 9-O-acetyl groups to that of sialic acid (SIA) in the monomer library of the CCP4 suite by using the program SKETCHER (CCP4 Suite).
Supplementary Material
Acknowledgments.
We thank Peter Rosenthal (MRC, Cambridge, UK) for providing coordinates of the HEF-inhibitor complex. We also thank P. Gros, J. de Groot-Mijnes, B. Bosch, P. Rottier, and P. Rosenthal for advice and for reading of this manuscript; G. Gerwig and J. Kamerling for help in preparing the sialic acid analogue; and P. Reeves (University of Essex, Colchester, UK) and M. Farzan (Harvard Medical School, MA) for sharing HEK293S GnTI(−) cells and expression plasmids, respectively. We acknowledge the European Synchrotron Radiation Facility for providing beamline facilities and the beamline scientists at ID-14-EH3 and ID23-1 for help with data collection. This work was supported by “Pionier” program and ECHO grants of the Council for Chemical Sciences of The Netherlands Organization for Scientific Research (NWO-CW) and by a grant of the Mizutani Foundation for Glycoscience.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.S.B. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 3CL4 (wild-type BCoV HE) and 3CL5 (BCoV HE0 complexed with Neu4,5,9Ac32Me)].
See Commentary on page 8807.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800502105/DCSupplemental.
References
- 1.Angata T, Varki A. Chemical diversity in the sialic acids and related alpha-keto acids: An evolutionary perspective. Chem Rev. 2002;102:439–469. doi: 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- 2.Rogers GN, Paulson JC. Receptor determinants of human and animal influenza virus isolates: Differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology. 1983;127:361–373. doi: 10.1016/0042-6822(83)90150-2. [DOI] [PubMed] [Google Scholar]
- 3.Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc Natl Acad Sci USA. 2004;101:4620–4624. doi: 10.1073/pnas.0308001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shinya K, et al. Avian flu: Influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 5.Wagner R, Matrosovich M, Klenk HD. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol. 2002;12:159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 6.Yuan P, et al. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure (London) 2005;13:803–815. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 7.de Groot RJ. Structure, function and evolution of the hemagglutinin-esterase proteins of corona- and toroviruses. Glycoconj J. 2006;23:59–72. doi: 10.1007/s10719-006-5438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palese P, Schulman JL, Bodo G, Meindl P. Inhibition of influenza and parainfluenza virus replication in tissue culture by 2-deoxy-2,3-dehydro-N-trifluoroacetylneuraminic acid (FANA) Virology. 1974;59:490–498. doi: 10.1016/0042-6822(74)90458-9. [DOI] [PubMed] [Google Scholar]
- 9.von Itzstein M, et al. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature. 1993;363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 10.Moscona A. Neuraminidase inhibitors for influenza. N Engl J Med. 2005;353:1363–1373. doi: 10.1056/NEJMra050740. [DOI] [PubMed] [Google Scholar]
- 11.Vijgen L, et al. Complete genomic sequence of human coronavirus OC43: Molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol. 2005;79:1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vlasak R, Luytjes W, Leider J, Spaan W, Palese P. The E3 protein of bovine coronavirus is a receptor-destroying enzyme with acetylesterase activity. J Virol. 1988;62:4686–4690. doi: 10.1128/jvi.62.12.4686-4690.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vlasak R, Luytjes W, Spaan W, Palese P. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc Natl Acad Sci USA. 1988;85:4526–4529. doi: 10.1073/pnas.85.12.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snijder EJ, den Boon JA, Horzinek MC, Spaan WJ. Comparison of the genome organization of toro- and coronaviruses: Evidence for two nonhomologous RNA recombination events during Berne virus evolution. Virology. 1991;180:448–452. doi: 10.1016/0042-6822(91)90056-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornelissen LA, et al. Hemagglutinin-esterase, a novel structural protein of torovirus. J Virol. 1997;71:5277–5286. doi: 10.1128/jvi.71.7.5277-5286.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smits SL, et al. Nidovirus sialate-O-acetylesterases: Evolution and substrate specificity of coronaviral and toroviral receptor-destroying enzymes. J Biol Chem. 2005;280:6933–6941. doi: 10.1074/jbc.M409683200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrler G, et al. The receptor-destroying enzyme of influenza C virus is neuraminate-O-acetylesterase. EMBO J. 1985;4:1503–1506. doi: 10.1002/j.1460-2075.1985.tb03809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hellebo A, Vilas U, Falk K, Vlasak R. Infectious salmon anemia virus specifically binds to and hydrolyzes 4-O-acetylated sialic acids. J Virol. 2004;78:3055–3062. doi: 10.1128/JVI.78.6.3055-3062.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vlasak R, Krystal M, Nacht M, Palese P. The influenza C virus glycoprotein (HE) exhibits receptor-binding (hemagglutinin) and receptor-destroying (esterase) activities. Virology. 1987;160:419–425. doi: 10.1016/0042-6822(87)90013-4. [DOI] [PubMed] [Google Scholar]
- 20.Herrler G, Durkop I, Becht H, Klenk HD. The glycoprotein of influenza C virus is the haemagglutinin, esterase and fusion factor. J Gen Virol. 1988;69:839–846. doi: 10.1099/0022-1317-69-4-839. [DOI] [PubMed] [Google Scholar]
- 21.Herrler G, Klenk HD. Structure and function of the HEF glycoprotein of influenza C virus. Adv Virus Res. 1991;40:213–234. doi: 10.1016/S0065-3527(08)60280-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenthal PB, et al. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature. 1998;396:92–96. doi: 10.1038/23974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luytjes W, Bredenbeek PJ, Noten AF, Horzinek MC, Spaan WJ. Sequence of mouse hepatitis virus A59 mRNA 2: Indications for RNA recombination between coronaviruses and influenza C virus. Virology. 1988;166:415–422. doi: 10.1016/0042-6822(88)90512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schultze B, Gross HJ, Brossmer R, Herrler G. The S protein of bovine coronavirus is a hemagglutinin recognizing 9-O-acetylated sialic acid as a receptor determinant. J Virol. 1991;65:6232–6237. doi: 10.1128/jvi.65.11.6232-6237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kazi L, Lissenberg A, Watson R, de Groot RJ, Weiss SR. Expression of hemagglutinin esterase protein from recombinant mouse hepatitis virus enhances neurovirulence. J Virol. 2005;79:15064–73. doi: 10.1128/JVI.79.24.15064-15073.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 27.Molgaard A, Kauppinen S, Larsen S. Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases. Structure (London) 2000;8:373–383. doi: 10.1016/s0969-2126(00)00118-0. [DOI] [PubMed] [Google Scholar]
- 28.Russell RJ, Stevens DJ, Haire LF, Gamblin SJ, Skehel JJ. Avian and human receptor binding by hemagglutinins of influenza A viruses. Glycoconj J. 2006;23:85–92. doi: 10.1007/s10719-006-5440-1. [DOI] [PubMed] [Google Scholar]
- 29.Lissenberg A, et al. Luxury at a cost? Recombinant mouse hepatitis viruses expressing the accessory hemagglutinin esterase protein display reduced fitness in vitro. J Virol. 2005;79:15054–63. doi: 10.1128/JVI.79.24.15054-15063.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klausegger A, et al. Identification of a coronavirus hemagglutinin-esterase with a substrate specificity different from those of influenza C virus and bovine coronavirus. J Virol. 1999;73:3737–3743. doi: 10.1128/jvi.73.5.3737-3743.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Regl G, et al. The hemagglutinin-esterase of mouse hepatitis virus strain S is a sialate-4-O-acetylesterase. J Virol. 1999;73:4721–4727. doi: 10.1128/jvi.73.6.4721-4727.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wurzer WJ, Obojes K, Vlasak R. The sialate-4-O-acetylesterases of coronaviruses related to mouse hepatitis virus: A proposal to reorganize group 2 Coronaviridae. J Gen Virol. 2002;83:395–402. doi: 10.1099/0022-1317-83-2-395. [DOI] [PubMed] [Google Scholar]
- 33.Li W, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: High-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci USA. 2002;99:13419–24. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamerling JP, Vliegenthart JFG. In: Clinical Biochemistry--Principles, Methods, Applications. Lawson AM, editor. Vol. 1. Berlin: Walter de Gruyter; 1989. pp. 176–263. [Google Scholar]
- 36.Wagaman PC, Spence HA, O'Callaghan RJ. Detection of influenza C virus by using an in situ esterase assay. J Clin Microbiol. 1989;27:832–836. doi: 10.1128/jcm.27.5.832-836.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 38.Collaborative Computational Project 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 39.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 40.Brunger AT, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 41.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.