Abstract
Activity-regulated gene expression is believed to play a key role in the development and refinement of neuronal circuitry. Nevertheless, the transcriptional networks that regulate synapse growth and plasticity remain largely uncharacterized. Here, we show that microRNA 132 (miR132) is an activity-dependent rapid response gene regulated by the cAMP response element-binding (CREB) protein pathway. Introduction of miR132 into hippocampal neurons enhanced dendrite morphogenesis whereas inhibition of miR132 by 2′O-methyl RNA antagonists blocked these effects. Furthermore, neuronal activity inhibited translation of p250GAP, a miR132 target, and siRNA-mediated knockdown of p250GAP mimicked miR132-induced dendrite growth. Experiments using dominant-interfering mutants suggested that Rac signaling is downstream of miR132 and p250GAP. We propose that the miR132–p250GAP pathway plays a key role in activity-dependent structural and functional plasticity.
Keywords: cAMP response element-binding (CREB) protein, transcription, CaM kinase, actin cytoskeleton, Rac
Neuronal activity regulates the development and modification of neuronal circuitry in part by activating genetic programs. Activity-regulated gene expression has been implicated in axon guidance, dendrite elaboration, synapse formation, and long-lasting synaptic plasticity (1, 2). Dendrites are the primary site of excitatory synapses, and their morphogenesis determines both the size and number of synaptic contacts (3). Although dendritic development is partly controlled by intrinsic factors, neuronal activity also plays a critical role. Indeed, the timing of afferent innervation and synapse formation coincides with the period of maximum growth and dendritic remodeling (3).
The transcription factor cAMP response element-binding (CREB) protein is a key regulator of dendritic growth (4) and activity-regulated dendritic refinement in mature neurons (5). Although CREB is believed to be a critical regulator of neuronal plasticity, few CREB targets have been directly linked to plasticity. To identify these genes, we developed a novel technology, termed serial analysis of chromatin occupancy (SACO) that facilitated the genome-wide identification of CREB target regions (6). We focused on microRNAs (miRNAs) because the ability of these molecules to repress gene expression is believed to play an important role in development, differentiation, proliferation, survival, and oncogenesis (7). Interestingly, a significant fraction of miRNAs are enriched or specifically expressed in the nervous system (8), and transcription of some miRNAs changes dynamically during brain development (9, 10). miRNAs have been implicated in development of neuronal asymmetry in Caenorhabditis elegans, maturation of sensory neurons in Drosophila, and neurite outgrowth and spine homeostasis in rodents (11–14). Although activity is believed to play an essential role in sculpting neuronal development, miRNAs induced by neuronal activity have not been described.
Here, we show that microRNA 132 (miR132) is an activity-regulated rapid response gene in cultured hippocampal neurons and intact mice. 2′O-methyl RNA inhibitors of miR132 attenuated activity-induced dendritic growth. Likewise, forced expression of miR132 in cultured neurons mimicked the effects of bicuculline on dendrite growth. We suggested (14) that miR132 regulates neuronal morphogenesis in developing neurons by repressing translation of the Rho family GTPase-activating protein, p250GAP. In this work, we show that neuronal activity also represses p250GAP translation in a miR132-dependent manner. Translational repression of p250GAP is essential because a p250GAP mutant containing an altered miR132 recognition element (MRE) attenuated activity-stimulated dendrite growth. We further show that the miR132-mediated repression of p250GAP and regulation of dendritic growth occur by modulating Rac family GTPases.
Results
Synaptic Activity Induces miR132 Expression.
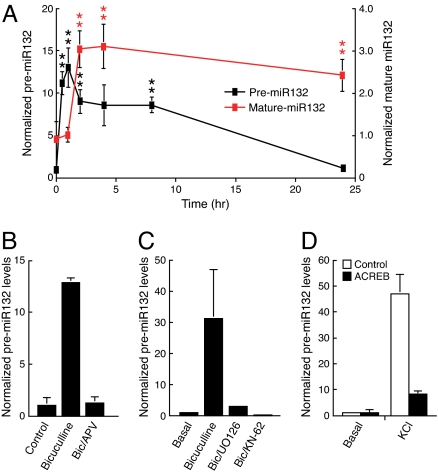
We showed (14) that BDNF treatment increased miR132 levels in immature neurons. In this work, we examined whether miR132 expression was also regulated by neuronal activity. Bicuculline-mediated inhibition of GABAA inhibitory tone increases spontaneous synaptic activity in our hippocampal neuron culture system [7 days in vitro (DIV 7)] (5). We used real-time PCR to quantify levels of the miR132 precursor transcript (premiR132) and mature miR132. Bicuculline triggered a rapid increase in expression of the miR132 precursor and mature miR132 (Fig. 1A). As expected, mature miR132 levels lagged induction of the precursor. Northern blot analysis confirmed the increase in miR132 levels in response to bicuculline stimulation [supporting information (SI) Fig. S1A]. In situ hybridization revealed a marked increase in miR132 levels in both soma and dendrites (Fig. S1B). This induction required activation of the NMDA receptor (NMDA-R) because pretreatment with the selective NMDA-R antagonist amino-5-phosphonovaleric acid (APV) attenuated the increase in miR132 expression (Fig. 1B). We recently demonstrated that synaptic activity regulated CREB-dependent transcription by activating the CaM kinase kinase (CaMKK), CaM kinase Iγ, and MEK-ERK pathway (5). We tested whether this pathway contributed to induction of miR132 by inhibiting CaM kinase with KN-62 and MEK-ERK with UO126. Both agents blocked miR132 induction by bicuculline (Fig. 1C).
Fig. 1.
miR132 is an activity- and CREB-regulated rapid response gene. (A) Hippocampal neurons were treated with 20 μM bicuculline for the indicated times. RNA was isolated, reverse-transcribed, and analyzed by real-time PCR with premiR132 cDNA primers (black squares) or mature-miR132 primers (red squares) by using the ABI TaqMan miRNA real-time PCR. The data were normalized to GAPDH cDNA levels also determined by real-time PCR (±SEM, n = 5–6). Error is SEM (**, P < 0.01). (B) Hippocampal neurons were treated with bicuculline for 1 h ±50 μM D-APV. PremiR132 levels were analyzed by real-time PCR and normalized to GAPDH. (C) Hippocampal neurons were pretreated with 10 μM UO126 or 10 μM KN-62 for 90 min and stimulated with 20 μM bicuculline ± inhibitors, as indicated, for 2 h. PremiR132 levels were analyzed by real-time PCR and normalized to GAPDH. (D) Hippocampal neurons were transfected with vector control or dominant-negative CREB (ACREB) and then treated 36 h later with KCl for 60 min. RNA was reverse-transcribed, analyzed by real-time PCR with premiR132 primers, and normalized to GAPDH cDNA levels.
Chromatin immunoprecipitation (ChIP) assays showed binding of CREB to the miR132 promoter in cultured hippocampal neurons and in the adult mouse hippocampus (Fig. S2). A negative control region in the GAPDH gene was not enriched, and no binding was detected in assays performed using a control antibody. We next examined whether activity regulates miR132 transcription via the CREB pathway by transfecting neurons with a CREB family inhibitor (ACREB) (15). KCl treatment increased transcription of the miR132 precursor (Fig. 1D), and ACREB expression attenuated this increase. These data suggest that neuronal activity regulates miR132 transcription predominantly via CREB.
miR132 Is Required for Activity-Dependent Dendritic Growth.
Recent studies have implicated the CREB pathway in depolarization-induced dendritic growth (4). Consequently, we examined whether miR132 might mediate this process. Dendritic morphology was measured by CAG promoter-driven expression of an EGFP-Map2B fusion gene. This fusion protein is selectively expressed in neurons and is largely restricted to dendrites (5). Dendritic length and branching were quantified on DIV 9. Quantitation for EGFP-MAP2 or endogenous MAP2 gave indistinguishable results (5). Bicuculline and miR132 increased total dendritic length by ≈70% and branching by ≈50% (Fig. 2 A–C). As a control, we showed that a nonneuronal miRNA, miR1-1, did not affect dendritic growth (data not shown). KCl caused a similar change in dendritic morphology (data not shown). Both bicuculline- and KCl-stimulated increases in dendritic length and branching were blocked by the NMDA receptor antagonists APV (5) and 7-Cl-kynurenic acid (Fig. S3A). Inhibition of NR2B-containing receptors with ifenprodil (Fig. S3B) or RO25-6981 (data not shown) also abolished bicuculline-stimulated growth. None of the treatments increased apoptosis, as measured by Hoechst staining or by quantitation of cleaved caspase 3 (data not shown).
Fig. 2.
Activity and CREB-regulated dendritic growth require miR132. (A–C) Hippocampal neurons were transfected with plasmids encoding MAP2B-EGFP ± miR132 or 2′OM-132 and treated with ±20 μM bicuculline for 48 h. Dendritic length (B) and branch number (C) were quantified at 9 DIV (***, P < 0.001). (D–F) Hippocampal neurons were transfected with plasmids encoding MAP2B-EGFP ± caCREB ± 2′OM-miR132. Dendritic length (E) and branch number (F) were quantified at 9 DIV.
We next examined whether miR132 is required for activity-dependent growth. Transfection of hippocampal neurons with a 2′O-methyl RNA oligonucleotide that targets miR132 (2′OM-132) blocked bicuculline-stimulated growth and branching (Fig. 2 A–C) but had no effect on basal dendritic length, indicating that miR132 specifically regulates activity-induced changes in dendritic morphology. Transfection of a constitutively active CREB mutant (caCREB) mimicked the bicuculline effects in a manner that was also blocked by the 2′OM-132 inhibitor (Fig. 2 D–F), suggesting that CREB regulates dendrite growth by controlling miR132 transcription.
p250GAP Is an Activity-Regulated miR132 Target.
In immature neurons, the Rho family GTPase-activating protein, p250GAP, is a miR132 target (14). Consequently, we sought to determine whether neuronal activity also regulated p250GAP via miR132. The putative miR132 MRE in p250GAP is flanked by two highly conserved hairpin structures. We cloned a region that flanked this conserved structure into the 3′-UTR of a luciferase reporter construct. The complement of the p250GAP MRE served as a negative control. The luciferase-MRE constructs were then expressed in hippocampal neurons with either empty vector (EV) or a vector expressing miR132. miR132 transfection markedly suppressed luciferase activity of the p250GAP MRE luciferase construct. In contrast, coexpression of miR132 with the reverse complement control had no effect on luciferase activity (Fig. 3A). We next generated a p250GAP mutant (mt-p250GAP) that contained a two-nucleotide deletion in the miR132 MRE seed sequence to test whether this site was required for miR132-mediated repression. Expression of miR132 attenuated expression of WT-p250GAP but had no effect on mt-p250GAP (Fig. 3B). Our finding that miR132 targets the p250GAP MRE in a heterologous reporter system and that an intact seed sequence is required for miR132-mediated repression suggests that p250GAP is a direct miR132 target. This finding was confirmed by showing that overexpression of miR132 in hippocampal neurons down-regulated endogenous p250GAP (Fig. 3C).
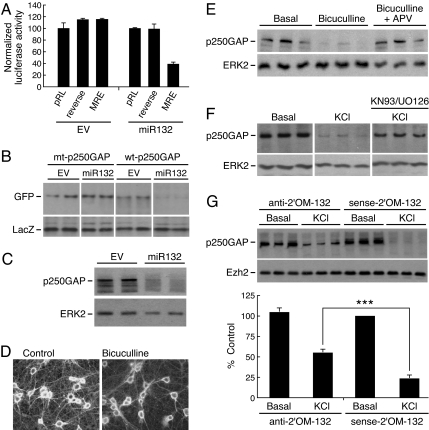
Fig. 3.
Synaptic activity regulates p250GAP via miR132. (A) The MRE in p250GAP is required for miR132 regulation. Hippocampal neurons were transfected with pRL-TK reporter plasmid (pRL) or a reporter containing the p250GAP MRE in the correct or reverse orientation. An excess of empty vector (EV) or miR132 plasmid was cotransfected. Renilla luciferase activity was normalized to cotransfected firefly luciferase activity. Data are expressed as percentage of wild-type pRL signal, and error is SEM. (B) miR132 inhibits p250GAP expression. Hippocampal neurons were transfected by electroporation with plasmids encoding either GFP-WT-p250GAP (wild type) or GFP-mt-p250GAP (miR132 MRE mutant) ± empty vector (EV) or miR132. After transfection, the neurons were cultured for 48 h. p250GAP expression was analyzed by Western blotting using an anti-GFP antibody. (C) MiR132 inhibits expression of p250GAP. Hippocampal neurons were transfected by Amaxa electroporation with either empty vector (EV) or plasmid encoding miR132. After transfection, the neurons were cultured for 48 h, and p250GAP expression was analyzed by Western blotting using an anti-p250GAP antibody. (D) Hippocampal neurons were treated with and without 20 μM bicuculline for 24 h and immunostained with anti-p250GAP. Representative images of control and stimulated neurons are shown. (E) Hippocampal neurons were treated ±20 μm bicuculline ± 50 μM D-APV for 24 h. Endogenous p250GAP and ERK2 expression was analyzed by Western blotting. (F) Hippocampal neurons were pretreated ±KN-93 and UO126 for 2 h, then stimulated with ±20 μM bicuculline for 24 h. Endogenous p250GAP and ERK2 expression was analyzed by Western blotting in triplicate samples. (G) Primary hippocampal DIV 7 neurons were treated with antisense miR132 antagomir or control sense antagomir for 24 h. Cells were then treated with KCl for 24 h and immunoblotted for endogenous p250GAP and ERK2. Quantitation of p250GAP levels in G is shown at the bottom of the figure (n = 6). Statistical analyses used ANOVA and Tukey's post test (±SEM, ***, P < 0.001).
We next examined whether synaptic activity repressed p250GAP expression and whether miR132 contributed to this regulation. Hippocampal neurons showed strong staining for p250GAP in the soma and dendrites. Bicuculline decreased p250GAP immunoreactivity in both compartments (Fig. 3D). Similarly, bicuculline decreased p250GAP protein levels in a manner that depended on NMDA-R signaling (Fig. 3E). Treatment of hippocampal neurons with the CaMK inhibitor, KN-93, and the MEK inhibitor, UO126, attenuated the decrease in p250GAP induced by KCl (Fig. 3F), suggesting that depolarization-mediated repression of p250GAP depends on CaMK-ERK-CREB signaling. Because p250GAP mRNA levels were not affected (data not shown), we propose that this change occurs at the level of translation.
To address whether miR132 was required for activity-dependent suppression of p250GAP, we used cholesterol-modified 2′O-methyl oligonucleotides (antagomirs) complementary to miR132 (anti-2′OM-132) or sense controls (sense-2′OM-132). Treatment with the antisense miR132 antagomir reduced the level of KCl-mediated repression of p250GAP (Fig. 3G). In contrast, the control sense antagomir had no effect, suggesting that down-regulation of p250GAP contributes to miR132-regulated dendritic growth.
Activity Regulates Dendritic Growth via miR132 Inhibition of p250GAP Translation.
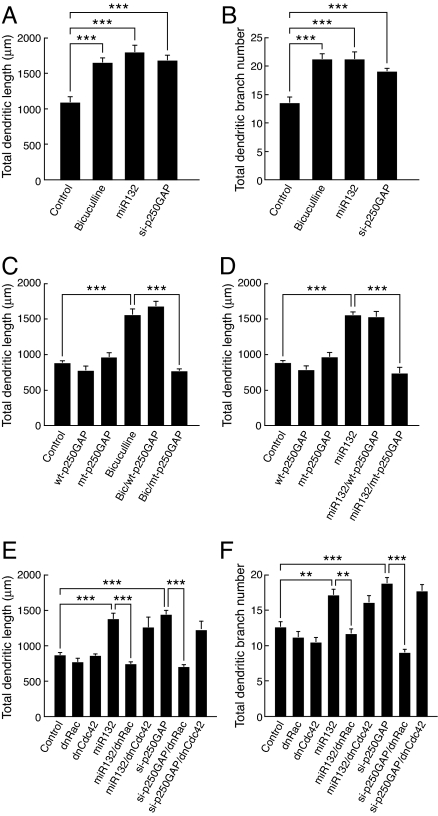
Next, we examined whether p250GAP repression contributes to activity-mediated dendritic growth. Transfection of an siRNA expression construct (si-p250GAP) that suppresses the expression of endogenous p250GAP (14) increased dendritic growth and branching by 50% (Fig. 4 A and B). Moreover, transfection of the miR132-insensitive p250GAP mutant (mt-p250GAP) attenuated activity- and miR132-dependent dendritic growth (Fig. 4 C and D). mt-p250GAP, but not the wild-type version, also blocked CREB-dependent dendritic growth (data not shown). Neither WT-p250GAP or mt-p250GAP affected basal dendritic length. These results led us to suggest that p250GAP down-regulation is important for bicuculline- and miR132-stimulated dendritic growth.
Fig. 4.
miR132 regulation of p250GAP is required for activity- and CREB-dependent dendritic growth. (A and B) Hippocampal neurons were transfected with plasmid encoding MAP2B-EGFP ± miR132 si-p250GAP (short-hairpin siRNA targeting p250GAP) and treated with ±20 μM bicuculline for 48 h. Dendritic length and branching were quantified at 9 DIV. (C and D) Expression of miR132-insensitive p250GAP (mt-p250GAP), but not wild-type p250GAP (WT-p250GAP), blocks bicuculline- and miR132-stimulated dendritic growth. Hippocampal neurons (7 DIV) were transfected with plasmids encoding MAP2B-EGFP ± either WT- or mt-p250GAP ± miR132 then treated with ±20 μM bicuculline. (E and F) Hippocampal neurons were transfected with plasmid encoding MAP2B-EGFP ± miR132 or si-p250GAP ± dnRac or dnCdc42 for 48 h. Dendritic growth was quantified at 9 DIV. Statistical analyses used ANOVA and Tukey's post test (±SEM, **, P < 0.01; ***, P < 0.001).
p250GAP has been reported to inhibit Rho family GTPases in vitro (16–20). To determine which Rho family GTPases participate in miR132- and p250GAP-regulated dendritic growth, we used dominant-interfering mutants. Expression of dominant-negative Rac (dnRac) and Cdc42 (dnCdc42) had little effect on basal dendritic growth or branching (Fig. 4 E and F). In contrast, dnRac largely blocked both miR132- and si-p250GAP-stimulated growth and branching. These data suggest that miR132 and p250GAP regulate dendritic growth by activating a Rac family GTPase.
Organotypic hippocampal slices retain the cellular and morphological organization of the intact hippocampus and have been used extensively to study aspects of hippocampal function (21, 22). We demonstrated that synaptic activity increases dendritic maturation in this preparation (5). Hippocampal slices from postnatal day 5 rat pups were cultured for 4 days and then treated with 20 μM bicuculline to mimic afferent input. Stimulation of cultured hippocampal slices with bicuculline increased premiR132 levels ≈5-fold over control slices (Fig. S4). Furthermore, the increase in premiR132 transcription was blocked by inhibition of the MEK-ERK and CaM kinase pathways.
We used biolistic transfection of a tomato fluorescent protein (TFP) marker to test whether miR132 is required for activity-dependent dendritic growth in the slice preparation (Fig. 5A). Both bicuculline and miR132 expression increased dendritic length by 2-fold (Fig. 5 A and B). A 2′O-methyl RNA oligonucleotide that targets miR132 had little effect under basal conditions but attenuated bicuculline-stimulated dendritic growth. siRNA-mediated knockdown of p250GAP also stimulated dendritic growth, suggesting that activity-dependent dendritic development in hippocampal slices similarly requires miR132 inhibition of p250GAP translation.
Fig. 5.
miR132 regulates dendritic morphology in organotypic hippocampal slices. (A) Hippocampal slices were cultured for 3 days, subjected to biolistic transfections with TFP ± other plasmids as indicated, allowed to recover for 1 day, then treated for 2 days with ±20 mM bicuculline. (B) Synaptic activity, miR132, and p250GAP regulate dendritic length of CA1 pyramidal neurons in slice culture. Representative examples of control or stimulated CA1 pyramidal neurons are depicted. Statistical analyses used ANOVA and Tukey's post test (±SEM, ***, P < 0.001).
Discussion
Activity-Dependent Regulation of miR132.
Activity-regulated gene expression is believed to regulate synapse clustering, synaptogenesis, developmental plasticity, synaptic plasticity, memory formation, addiction, the biological clock, and other behavioral adaptations. Recent genomic screens suggest the existence of thousands of novel noncoding RNAs, the majority of which are not represented on conventional microarrays (23, 24). A role for noncoding transcription in activity-regulated plasticity has not been established. In this work, we show that the noncoding RNA, miR132, is rapidly induced by neuronal activity. Moreover, we delineate an activity-regulated miRNA pathway that regulates dendritic morphogenesis by inhibiting translation of the synaptic protein, p250GAP. We focused on p250GAP because it was the only predicted miR132 target that showed perfect conservation across the vertebrate phylum. In vivo pull-down assays suggest that p250GAP potentially regulates several Rho family GTPases (16–20), and cerebellar granule cells from p250GAP-knockout mice show increased Cdc42 activity (18). p250GAP is enriched in the postsynaptic density where it interacts with the NMDA NR2B receptor subunit and the scaffold protein PSD-95 (25, 26). p250GAP also interacts with Fyn (17), a tyrosine kinase that phosphorylates NR2B and regulates NMDA-dependent neuronal plasticity (27). Interestingly, p250GAP was shown to interact with β-catenin (26), another regulator of synapse formation and dendrite growth (28, 29). Collectively, these studies show that p250GAP interacts with multiple synapse-specific proteins.
In this work, we show that neuronal activity triggers suppression of p250GAP levels in hippocampal neurons. The regulation of p250GAP levels by neuronal activity is markedly attenuated by selective inhibition of the miR132 pathway. The lack of a complete block by miR132 inhibitors may indicate that other pathways regulate p250GAP levels as well. The ability of miR132 to repress translation of exogenous p250GAP requires an intact miR132MRE, suggesting that miR132 plays a major role in activity-dependent regulation of p250GAP. CaM kinase II phosphorylates p250GAP in vitro and inhibits its GTPase-activating protein function (19). Intriguingly, the localization of p250GAP at the postsynaptic density may also be regulated by NMDA receptor signaling (19). These studies raise the possibility that NMDA receptor-dependent activity could also regulate p250GAP. We suggest that down-regulation of p250GAP function in dendrites is a critical mechanism by which neuronal activity modulates structural plasticity.
By suppressing p250GAP levels, miR132 expression presumably results in prolonged localized increases in Rac activity. Interestingly, other regulators of dendrite and spine growth, such as EphB, Kalirinin, and Tiam1, also show selectivity for Rac in hippocampal neurons (30, 31). Several downstream effectors of Rac and Cdc42, including Pak, Lim-kinase, and myosin heavy chain IIb, have been proposed to regulate structural or functional dendritic plasticity (32). Thus, we propose that miR132 regulates dendrite growth by down-regulating p250GAP and increasing Rac activity. This pathway may also contribute to activity-regulated actin remodeling.
Neuronal activity and calcium signaling play critical roles in dendritic development and plasticity (1, 3). In some models of neuronal plasticity, de novo gene expression is believed to be required for changes in synapse structure (33). In particular, the CREB transcriptional pathway has been implicated in structural plasticity associated with long-term facilitation (34). Inhibition of the CREB pathway reduces dendrite growth in cortical neurons (4), and CREB activity is necessary and sufficient for activity-regulated dendritic plasticity (5). In this work, we provide evidence that CREB regulates dendrite arborization via miR132. Our observation that a p250GAP MRE mutant attenuates activity- and CREB-mediated dendritic outgrowth supports an essential role for the CREB–miR132 pathway in morphological plasticity. The CREB-regulated trophic factor Wnt2 also regulates activity-regulated dendritic outgrowth. Because p250GAP interacts with the Wnt effector, β-catenin, it is possible that Wnt2 also cooperates with the miR132 pathway. Because transcriptional repression is often associated with long-lasting chromatin remodeling, one of the functions of miRNA rapid response genes might be to facilitate reversible repression of gene expression.
Components of the miRNA RISC complex are enriched in dendrites, which suggests that miRNAs might be well positioned to repress gene expression in this cellular compartment. In support of this idea, a recent study showed that miR134 and its target, LIMK, were colocalized in dendrites (13). Recent studies indicate that formation of functional miRNA–RISC complexes might also be regulated by cellular signaling (35), and it is conceivable that miRNA function could be regulated by signals limited to the dendritic or synaptic compartment. Although our data indicate that miR132 regulates morphogenesis by repressing translation of p250GAP, we recently showed that MeCP2 is also a miR132 target (36). Interestingly, MeCP2 has also been proposed to regulate dendritic growth. Future experiments will address the role of the miR132–MeCP2 pathway in dendrite growth and patterning. Bioinformatic algorithms predict other conserved targets that could also contribute to miR132 function. The ability of miRNAs to repress targets in a combinatorial manner suggests that these molecules could function as neuronal coincidence detectors and contribute in additional ways to synaptic plasticity.
Materials and Methods
Reagents and Plasmids.
The following reagents were purchased from the indicated sources: UO126 (Calbiochem); STO-609, NMDA, and APV (Tocris Cookson). The CREB antibody was described in ref. 37. Antibodies were also obtained from the following sources: Erk2 and Ezh2 (Santa Cruz Biotechnology), anti-GFP (Clontech), and anti-LacZ (Promega). Map2B-GFP (5) and caCREB (38) plasmids have been described previously. The pCAG-ACREB, pCAG-LacZ, CRE-luciferase (39), and pCAG-miR132 (14) plasmids were also described previously. The p250GAP miR132 MRE sequence was amplified from a human cDNA with primers GCCCCGGGAGCAATAGAGTT and TGGGAGGGGAAGGTGGTGAT and cloned into the XbaI site of pRL-TK (Promega). The p250GAP mutant was generated by PCR-based mutagenesis of GFP-tagged p250GAP (19) with the following primers: GGTTATTGAAAAAAATAGAAGTCCACTGTCCAGCAGAGG and CCTCTGCTGGACAGTGGACTTCTATTTTTTTCAATAACC. The sequences of the 2′O-methyl oligoribonucleotides (IDT) are: antisense, GGGCGACCAUGGCUGUAGACUGUUACUGUGG; sense, UCCAUUGUCAGAUGUCGGUACCAGCGGGGCG. The 2′O-methyl antagomir oligoribonucleotides sequences are: CGACCAUGGCUGUAGACUGUUA-3′chol, miR132; and UAACAGUCUACAGCCAUGGUCG-3′chol, sense.
Cell Culture.
Hippocampal neurons (2 × 105 cells per square centimeter) were cultured from P1-2 Sprague–Dawley rats on plates coated with poly-l-lysine (Sigma; molecular weight 300,000) as described in ref. 5. Hippocampal neurons were maintained in Neurobasal A medium (Invitrogen) supplemented with B27 (Invitrogen), 0.5 mM l-glutamine, and 5 μM cytosine-d-arabinofuranoside (Sigma; added at 2 DIV). Hippocampal neurons were then cultured a further 3–7 days, at which time they were either transfected or treated with various pharmacological reagents as specified in the text or figure legends.
Transfection.
Primary hippocampal neurons were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocols. In each experiment, we optimized DNA amounts, transfection reagent amounts, and transfection duration to minimize toxicity and maximize transfection efficiency. None of the transfections or drug treatments had noticeable effects on apoptosis as assessed by condensed nuclei using Hoechst staining. Luciferase and β-galactosidase activities were measured by using luciferase and Galacoto-Light assay kits (Perkin–Elmer).
Quantification of Dendritic Morphology.
High-density hippocampal neurons were transfected on 5–6 DIV and were fixed (4% paraformaldehyde, 3% sucrose, 60 mM Pipes, 25 mM Hepes, 5 mM EGTA, 1 mM MgCl2, pH7.4) 4 days later for 20 min at room temperature. Fluorescent images were acquired by using a cooled CCD camera (Hamamatsu Photonics) attached to a Zeiss Axioplan2 (Carl Zeiss) inverted microscope with a 63× oil-immersion lens. Morphometric measurements were performed by using Openlab software (Improvision). Dendrites were analyzed as described in ref. 5. Each experiment was repeated at least three times with independent neuronal preparations.
Slice Culture and Transfection.
Organotypic hippocampal slices from P5 Sprague–Dawley rats were cultured for 3 days as described in ref. 5. To visualize dendritic arbors, slices were transfected with pCAG-TFP (six test plasmids) by using a Helios gene gun (Bio-Rad), according to the manufacturer's protocol. After transfection, slices were allowed to recover for 24 h before stimulation with 16 mM KCl or 20 mM bicuculline-methiodide (Tocris) for 2 days. Slices were fixed, mounted, and imaged with a confocal microscope. Dendritic processes were measured as described above.
Western Blotting and ChIP.
Western blotting was conducted as described in ref. 39. The following primary antibodies were used overnight at 4°C in Tris-buffered saline containing 0.1% Triton X-100, 10 mM NaF, and 5% BSA: polyclonal anti-p250GAP (1:1,000) (19), monoclonal anti-GFP (Clontech), polyclonal anti-Ezh2 (Santa Cruz Biotechnology), and monoclonal anti-LacZ (Promega). Neonatal 7 DIV rat hippocampal neurons (2 × 106) were subjected to chromatin immunoprecipitation as described in ref. 6. For chromatin immunoprecipitation from 8-week mouse hippocampus, tissue was minced into ≈1-mm cubes and fixed with 4% paraformaldehyde in PBS for 30 min at 4°C. The fixed tissue was then processed as for cultured neurons.
Reverse Transcription.
Neurons were treated as described, and total RNA was isolated by using TRIzol (Invitrogen) according to the manufacturer's instructions. RNA (50 ng to 1 μg) was reverse transcribed with Moloney murine leukemia virus reverse transcriptase (Invitrogen) and 50–250 ng of random primers (Invitrogen).
Quantitative Real-Time PCR.
PCRs (10 μl) contained 1 μl of 10× PCR buffer (Invitrogen), 2.5 mM MgCl2, 200 mM dNTP (Roche), 0.125–0.25 mM primer (IDT), 13 SYBR Green I (Invitrogen), and 1 unit of platinum Taq (Invitrogen). All RT-PCR was run on an Opticon OP346 (MJ Research) for one cycle at 95°C for 35 s, and 30–50 cycles at 94°C for 15 s, and 68°C for 40 s. CREB ChIPs were expressed as nanograms of purified input genomic DNA. The real-time PCR was analyzed by using the linear standard curve method. All standard curves had an R2 of at least 0.995, were composed of a minimum of 5 points, and were linear for at least 3 orders of magnitude. To avoid plateau effects, the Ct was always positioned in the logarithmic component of the sigmoid fluorescence curve. The Ct was selected based solely on the maximal linearity of standard curve. RT-PCR data were normalized to GAPDH cDNA levels also detected by real-time PCR (other housekeeping genes showed similar results). All RT-PCR data showed at least 100-fold higher levels of product than no reverse transcriptase controls. The following primers were used: RT miR132-precursor-1 CCTCCGGTTCCCACAGTAACAA, RT miR132-precursor-2 CCGCGTCTCCAGGGCAAC, GAPDH-1 AGTGCCAGCCTCGTCCCGTAG, GAPDH-2 CCAAATCCGTTCACACCGACCTT, ChIP miR132-57-1 CACGCTCCCCACCACTCC, ChIP miR132-57-2 TTGCTCTGTATCTGCCCAAACC. Real-time PCR for mature miR132 was performed by using TaqMan microRNA assay (Applied Biosystems) according to the manufacturer's protocol.
Immunocytochemistry.
Hippocampal neurons were fixed in 4% paraformaldehyde, 4% sucrose, PBS, and 50 mM Hepes (pH 7.5) at 37°C for 15 min. The cells were rinsed three times for 5 min in PBS, permeabilized with 0.2% Triton X-100 in PBS for 5 min, and rinsed three times for 5 min in blocking buffer [PBS containing 0.5% fish gelatin (Sigma), 0.05% Tween 20]. The cells were then stained with anti-p250GAP (1:10,000) in blocking buffer for 60 min at room temperature and washed three times for 5 min in blocking buffer. Coverslips were mounted on glass slides and analyzed by fluorescence microscopy as described above.
In Situ Hybridization.
The following fluorescein-labeled miRNA probes were obtained from IDT: miR132 AS 5′-CGACCAUGGCUGUAGACUGUUA-FAM-3′, miR132 scrambled 5′-UAACAGUCUACAGCCAUGGUCG-FAM-3′. Hybridization was performed as described in ref. 40 with the following exceptions. A hybridization solution consisting of probe (300 ng/ml) and tRNA (1.9 mg/ml) was applied to coverslips, placed in a humid chamber, and incubated overnight at 34°C. The following day, coverslips were rinsed as described in the Exiqon protocol for miRNA in situ hybridization of frozen tissue sections (Exiqon). Coverslips were stained by using the Alexa Fluor 488 signal-amplification kit for fluorescein (Invitrogen) and mounted on slides by using Vectashield Hard Set (Vector Laboratories).
Statistical Analyses.
Raw data approximated normality (the Shapiro–Wilk test) and the assumption of equal variances (Levene's test). ANOVA and Tukey's post test were used to test the null hypothesis.
Supplementary Material
Acknowledgments.
We thank Jami Dwyer and Amir Bashar for technical assistance and Gail Mandel for comments and suggestions. This work was supported by grants from the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803072105/DCSupplemental.
References
- 1.Katz LC, Shatz CJ. Synaptic activity and the construction of cortical circuits. Science. 1996;274:1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- 2.Yuste R, Bonhoeffer T. Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annu Rev Neurosci. 2001;24:1071–1089. doi: 10.1146/annurev.neuro.24.1.1071. [DOI] [PubMed] [Google Scholar]
- 3.Wong RO, Ghosh A. Activity-dependent regulation of dendritic growth and patterning. Nat Rev Neurosci. 2002;3:803–812. doi: 10.1038/nrn941. [DOI] [PubMed] [Google Scholar]
- 4.Redmond L, Kashani AH, Ghosh A. Calcium regulation of dendritic growth via CaM kinase IV and CREB-mediated transcription. Neuron. 2002;34:999–1010. doi: 10.1016/s0896-6273(02)00737-7. [DOI] [PubMed] [Google Scholar]
- 5.Wayman GA, et al. Activity-dependent dendritic arborization mediated by CaM-kinase I activation and enhanced CREB-dependent transcription of Wnt-2. Neuron. 2006;50:897–909. doi: 10.1016/j.neuron.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Impey S, et al. Defining the CREB regulon: A genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 7.Du T, Zamore PD. MicroPrimer: The biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 8.Klein ME, Impey S, Goodman RH. Role reversal: The regulation of neuronal gene expression by microRNAs. Curr Opin Neurobiol. 2005;15:507–513. doi: 10.1016/j.conb.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 9.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miska EA, et al. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schratt GM, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 14.Vo N, et al. A cAMP-response element-binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci USA. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahn S, et al. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol Cell Biol. 1998;18:967–977. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao C, et al. GC-GAP, a Rho family GTPase-activating protein that interacts with signaling adapters Gab1 and Gab2. J Biol Chem. 2003;278:34641–34653. doi: 10.1074/jbc.M304594200. [DOI] [PubMed] [Google Scholar]
- 17.Taniguchi S, et al. p250GAP, a neural RhoGAP protein, is associated with and phosphorylated by Fyn. Biochem Biophys Res Commun. 2003;306:151–155. doi: 10.1016/s0006-291x(03)00923-9. [DOI] [PubMed] [Google Scholar]
- 18.Nasu-Nishimura Y, et al. Role of the Rho GTPase-activating protein RICS in neurite outgrowth. Genes Cells. 2006;11:607–614. doi: 10.1111/j.1365-2443.2006.00966.x. [DOI] [PubMed] [Google Scholar]
- 19.Nakazawa T, et al. p250GAP, a novel brain-enriched GTPase-activating protein for Rho family GTPases, is involved in the N-methyl-d-aspartate receptor signaling. Mol Biol Cell. 2003;14:2921–2934. doi: 10.1091/mbc.E02-09-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moon SY, Zang H, Zheng Y. Characterization of a brain-specific Rho GTPase-activating protein, p200RhoGAP. J Biol Chem. 2003;278:4151–4159. doi: 10.1074/jbc.M207789200. [DOI] [PubMed] [Google Scholar]
- 21.Bahr BA. Long-term hippocampal slices: A model system for investigating synaptic mechanisms and pathologic processes. J Neurosci Res. 1995;42:294–305. doi: 10.1002/jnr.490420303. [DOI] [PubMed] [Google Scholar]
- 22.Caeser M, Aertsen A. Morphological organization of rat hippocampal slice cultures. J Comp Neurol. 1991;307:87–106. doi: 10.1002/cne.903070109. [DOI] [PubMed] [Google Scholar]
- 23.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 24.Cheng J, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 25.Nakagawa S, et al. Localization of phosphorylated cAMP response element-binding protein in immature neurons of adult hippocampus. J Neurosci. 2002;22:9868–9876. doi: 10.1523/JNEUROSCI.22-22-09868.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okabe T, et al. RICS, a novel GTPase-activating protein for Cdc42 and Rac1, is involved in the β-catenin–N-cadherin and N-methyl-d-aspartate receptor signaling. J Biol Chem. 2003;278:9920–9927. doi: 10.1074/jbc.M208872200. [DOI] [PubMed] [Google Scholar]
- 27.Grant SG, et al. Impaired long-term potentiation, spatial learning, and hippocampal development in fyn mutant mice. Science. 1992;258:1903–1910. doi: 10.1126/science.1361685. [DOI] [PubMed] [Google Scholar]
- 28.Murase S, Mosser E, Schuman EM. Depolarization drives β-catenin into neuronal spines promoting changes in synaptic structure and function. Neuron. 2002;35:91–105. doi: 10.1016/s0896-6273(02)00764-x. [DOI] [PubMed] [Google Scholar]
- 29.Yu X, Malenka RC. β-Catenin is critical for dendritic morphogenesis. Nat Neurosci. 2003;6:1169–1177. doi: 10.1038/nn1132. [DOI] [PubMed] [Google Scholar]
- 30.Penzes P, et al. Rapid induction of dendritic spine morphogenesis by trans-synaptic ephrinB–EphB receptor activation of the Rho-GEF kalirin. Neuron. 2003;37:263–274. doi: 10.1016/s0896-6273(02)01168-6. [DOI] [PubMed] [Google Scholar]
- 31.Tolias KF, et al. The Rac1-GEF Tiam1 couples the NMDA receptor to the activity-dependent development of dendritic arbors and spines. Neuron. 2005;45:525–538. doi: 10.1016/j.neuron.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 32.Van Aelst L, Cline HT. Rho GTPases and activity-dependent dendrite development. Curr Opin Neurobiol. 2004;14:297–304. doi: 10.1016/j.conb.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 33.Bailey CH, Montarolo P, Chen M, Kandel ER, Schacher S. Inhibitors of protein and RNA synthesis block structural changes that accompany long-term heterosynaptic plasticity in aplysia. Neuron. 1992;9:749–758. doi: 10.1016/0896-6273(92)90037-e. [DOI] [PubMed] [Google Scholar]
- 34.Bartsch D, Casadio A, Karl KA, Serodio P, Kandel ER. CREB1 encodes a nuclear activator, a repressor, and a cytoplasmic modulator that form a regulatory unit critical for long-term facilitation. Cell. 1998;95:211–223. doi: 10.1016/s0092-8674(00)81752-3. [DOI] [PubMed] [Google Scholar]
- 35.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 36.Klein ME, et al. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10:1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- 37.Moreno CS, Beresford GW, Louis-Plence P, Morris AC, Boss JM. CREB regulates MHC class II expression in a CIITA-dependent manner. Immunity. 1999;10:143–151. doi: 10.1016/s1074-7613(00)80015-1. [DOI] [PubMed] [Google Scholar]
- 38.Cardinaux JR, et al. Recruitment of CREB binding protein is sufficient for CREB-mediated gene activation. Mol Cell Biol. 2000;20:1546–1552. doi: 10.1128/mcb.20.5.1546-1552.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arthur JS, et al. Mitogen- and stress-activated protein kinase 1 mediates cAMP response element-binding protein phosphorylation and activation by neurotrophins. J Neurosci. 2004;24:4324–4332. doi: 10.1523/JNEUROSCI.5227-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cunningham MJ, Scarlett JM, Steiner RA. Cloning and distribution of galanin-like peptide mRNA in the hypothalamus and pituitary of the macaque. Endocrinology. 2002;143:755–763. doi: 10.1210/endo.143.3.8661. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.