Abstract
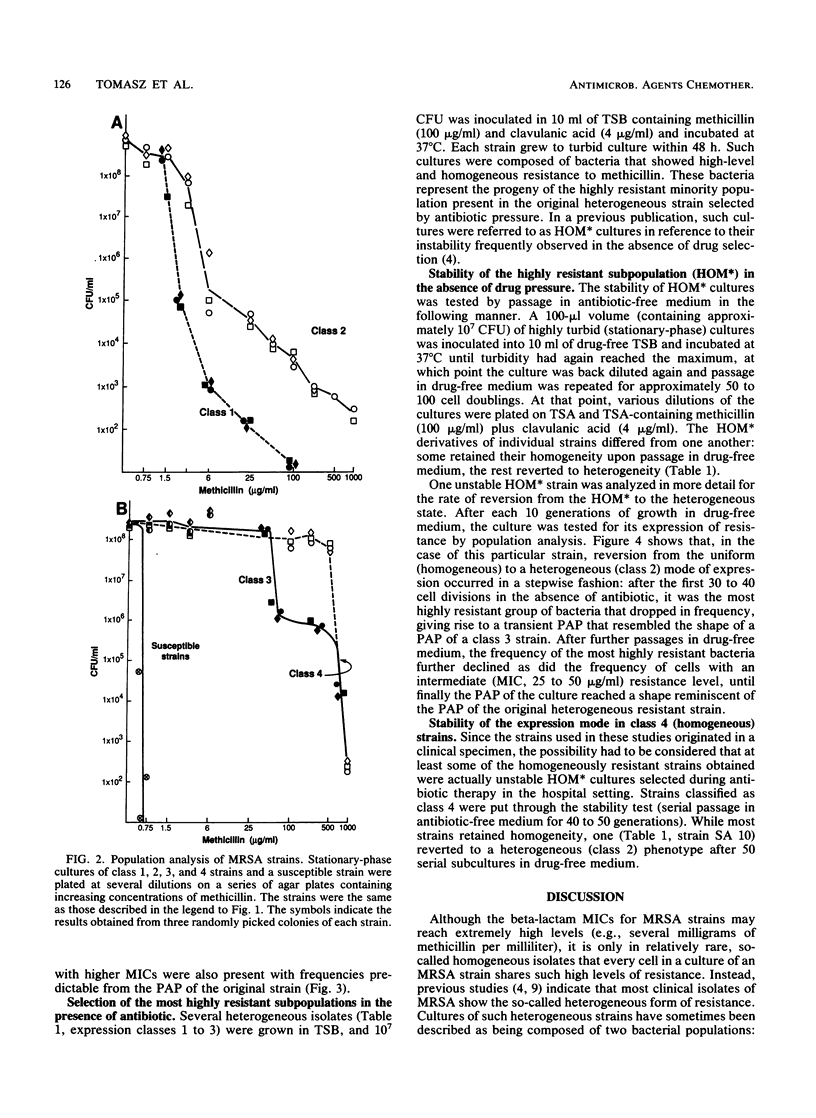
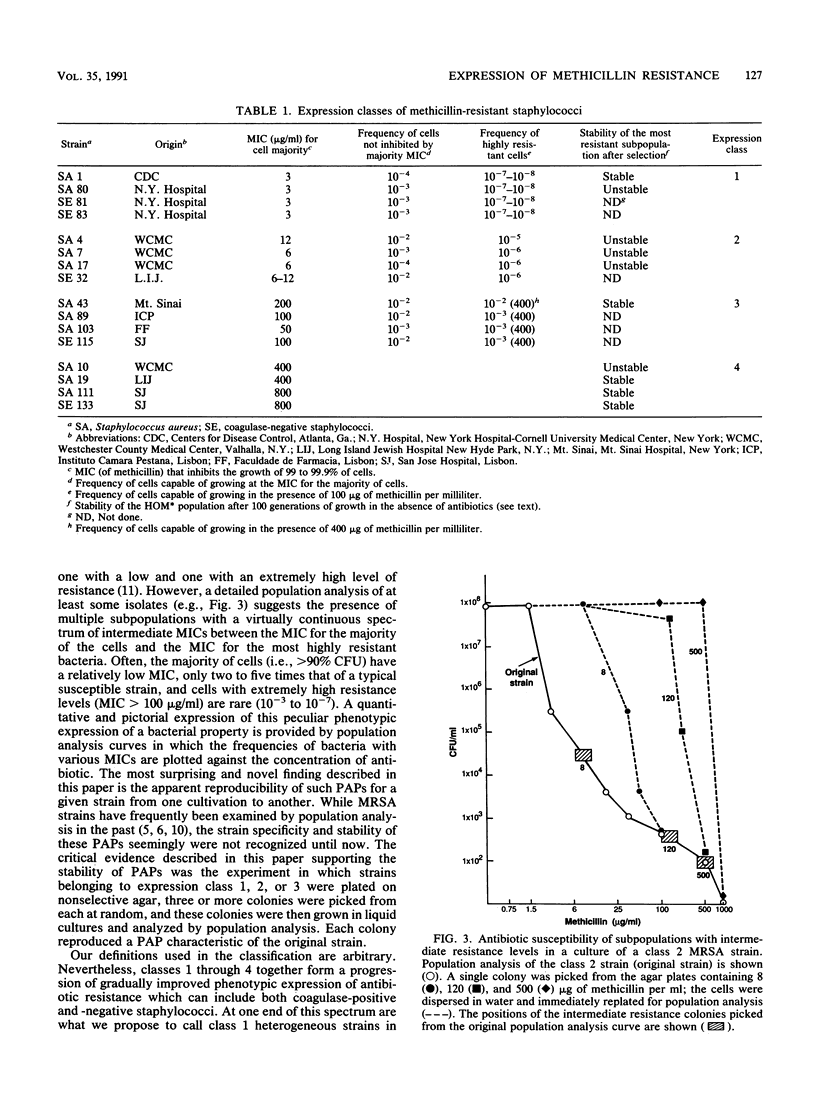
A collection of coagulase-positive and -negative clinical strains of staphylococci, all of which gave a positive reaction with a mec-specific DNA probe, was analyzed for the mode of phenotypic expression of methicillin resistance by using population analysis on agar plates containing different concentrations of the antibiotic. Strains could be divided into four arbitrary expression classes. Cultures of class 4 strains were composed of uniformly and highly resistant bacteria (MIC greater than or equal to 800 micrograms/ml). In contrast, cultures of strains belonging to classes 1, 2, and 3 were heterogeneous: they were composed of two or more subpopulations of cells that differed from one another in MICs and frequencies. In cultures of strains belonging to expression class 1, most of the cells had methicillin MICs of 1.5 to 3 micrograms/ml, i.e., only two to three times higher than those for truly susceptible strains. In cultures of strains belonging to expression classes 2 and 3, the methicillin MICs for the majority of bacteria ranged from 6 to 12 and up to 50 to 200 micrograms/ml, respectively. While the definition of the expression classes was arbitrary, the modes of phenotypic expression were specific and reproducible: randomly picked colonies of a given strain produced identical population profiles. The strain-specific mode of expression was also retained after numerous single-colony picks and sequential passages in antibiotic-free medium. We suggest that these classes represent stages in an evolutionary sequence leading to progressively improved phenotypic expression of methicillin resistance in staphylococci.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berger-Bächi B., Barberis-Maino L., Strässle A., Kayser F. H. FemA, a host-mediated factor essential for methicillin resistance in Staphylococcus aureus: molecular cloning and characterization. Mol Gen Genet. 1989 Oct;219(1-2):263–269. doi: 10.1007/BF00261186. [DOI] [PubMed] [Google Scholar]
- Hackbarth C. J., Chambers H. F. Methicillin-resistant staphylococci: detection methods and treatment of infections. Antimicrob Agents Chemother. 1989 Jul;33(7):995–999. doi: 10.1128/aac.33.7.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman B. J., Tomasz A. Expression of methicillin resistance in heterogeneous strains of Staphylococcus aureus. Antimicrob Agents Chemother. 1986 Jan;29(1):85–92. doi: 10.1128/aac.29.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneine N., Stewart P. R. Physiological determination of methicillin resistance in Staphylococcus aureus: comparison of clinical and genetically derived isolates. J Antimicrob Chemother. 1986 Jun;17(6):705–715. doi: 10.1093/jac/17.6.705. [DOI] [PubMed] [Google Scholar]
- Inglis B., Matthews P. R., Stewart P. R. The expression in Staphylococcus aureus of cloned DNA encoding methicillin resistance. J Gen Microbiol. 1988 Jun;134(6):1465–1469. doi: 10.1099/00221287-134-6-1465. [DOI] [PubMed] [Google Scholar]
- Murakami K., Tomasz A. Involvement of multiple genetic determinants in high-level methicillin resistance in Staphylococcus aureus. J Bacteriol. 1989 Feb;171(2):874–879. doi: 10.1128/jb.171.2.874-879.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabath L. D. Chemical and physical factors influencing methicillin resistance of Staphylococcus aureus and Staphylococcus epidermidis. J Antimicrob Chemother. 1977 Nov;3 (Suppl 100):47–51. doi: 10.1093/jac/3.suppl_c.47. [DOI] [PubMed] [Google Scholar]
- Sabath L. D., Wallace S. J. The problems of drug-resistant pathogenic bacteria. Factors influencing methicillin resistance in staphylococci. Ann N Y Acad Sci. 1971 Jun 11;182:258–266. doi: 10.1111/j.1749-6632.1971.tb30662.x. [DOI] [PubMed] [Google Scholar]
- Thornsberry C., McDougal L. K. Successful use of broth microdilution in susceptibility tests for methicillin-resistant (heteroresistant) staphylococci. J Clin Microbiol. 1983 Nov;18(5):1084–1091. doi: 10.1128/jcm.18.5.1084-1091.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasz A., Drugeon H. B., de Lencastre H. M., Jabes D., McDougall L., Bille J. New mechanism for methicillin resistance in Staphylococcus aureus: clinical isolates that lack the PBP 2a gene and contain normal penicillin-binding proteins with modified penicillin-binding capacity. Antimicrob Agents Chemother. 1989 Nov;33(11):1869–1874. doi: 10.1128/aac.33.11.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]




