Abstract
Recently, cryoelectron microscopy of isolated macromolecular complexes has advanced to resolutions below 10 Å, enabling direct visualization of α-helical secondary structure. To help correlate such density maps with the amino acid sequences of the component proteins, we advocate peptide-based difference mapping, i.e., insertion of peptides, ≈10 residues long, at targeted points in the sequence and visualization of these peptides as bulk labels in cryoelectron microscopy-derived difference maps. As proof of principle, we have appended an extraneous octapeptide at the N terminus of hepatitis B virus capsid protein and determined its location on the capsid surface by difference imaging at 11 Å resolution. Hepatitis B virus capsids are icosahedral particles, ≈300 Å in diameter, made up of T-shaped dimers (subunit Mr, 16–21 kDa, depending on construct). The stems of the Ts protrude outward as spikes, whereas the crosspieces pack to form the contiguous shell. The two N termini per dimer reside on either side of the spike-stem, at the level at which it enters the shell. This location is consistent with formation of the known intramolecular disulfide bond between the cysteines at positions 61 and −7 (in the residual propeptide) in the “e-antigen” form of the capsid protein and has implications for why this clinically important antigen remains unassembled in vivo.
Keywords: hepatitis B virus core antigen/virus capsid structure/three-dimensional image reconstruction
Cryoelectron microscopy (cryo-EM) of macromolecular particles recently has achieved resolutions below 10 Å (1–3) and holds great promise for the analysis of the large complexes that control many fundamental biological processes and whose structures appear likely, in many cases, to remain beyond the reach of x-ray crystallography. A high-priority goal is to develop ways to delineate the overall paths of polypeptide chains through density maps in the 7 Å to 10 Å resolution range. Several approaches have been proposed, all of which involve modifying the specimen in some chemically defined way and visualizing the concomitant structural change by cryo-EM, i.e., variations on the theme of difference mapping. Viable modifications include labeling with heavy metal clusters (4, 5) or binding antibodies against defined linear epitopes (6, 7). We have now explored the potentialities of appending small peptides (≈10 residues) to serve as bulk labels and report localization of the N terminus of hepatitis B virus (HBV) capsid protein by this approach.
HBV, a major human pathogen (8), is a small enveloped virus whose limited genetic coding capacity is enhanced by expression of the same gene from different initiation sites (9). Thus, there are two variants of the capsid protein, HBcAg (core antigen) and HBeAg (e-antigen). The HBcAg capsid plays a crucial role in the viral replication cycle. HBeAg, on the other hand, is secreted in unassembled form and, although it is a clinically useful marker for the state of infection, its biological role remains obscure. Both proteins contain the 149-residue assembly domain but their termini differ (Fig. 1). HBcAg has a 34-residue RNA-binding domain at its C-terminus that is absent from HBeAg, which, unlike HBcAg, retains 10 residues of a 29-residue N-terminal propeptide.
Figure 1.
Comparison of the N- and C-terminal sequences of four variants of the HBV capsid protein. In its N terminus (residues −8 to 2), Cpe has a 10-residue extension derived from β-galactosidase and lacks the first two residues of the wild-type assembly domain; also, it lacks the five distal residues of the wild-type assembly domain, but has two extraneous C-terminal residues (Ile 145, Ser 146). The reference construct, Cp147, has a normal N terminus and terminates at position 147. The 34-residue “protamine” domain at the C terminus of HBcAg engages in RNA binding.
In vivo, HBcAg self-assembles into capsids enclosing the viral RNA pregenome and reverse transcriptase (10). In vitro (11) and in heterologous expression systems (12, 13), the assembly domain suffices for production of icosahedral capsids. These capsids are of two sizes, with triangulation numbers (14) of T = 3 (90 dimers, diameter ≈290 Å) and T = 4 (120 dimers, diameter ≈330 Å), respectively (15, 11). Both termini of the assembly domain influence the products obtained. With a full-length assembly domain, 90–95% of capsids are T = 4, but truncation from the C terminus progressively increases the proportion of T = 3 capsids (16) until 11 residues are removed, abolishing assembly competence (17). Deletions of up to nine residues from the N terminus are tolerated (ref. 18; S.S. and P.W., unpublished results), as are some sizable N-terminal extensions (19, 20).
Because the assembly domain is highly α-helical (11), much of its secondary structure was visualized at resolutions of 9 Å (2) and 7.4 Å (1). Thus, subunits were seen to pair into dimers, the building-blocks for capsid assembly (21), by formation of a four-helix bundle from two helix–loop–helix motifs (1, 2). Specific proposals also have been made for much of the rest of the fold (1, 22). The N terminus represents an important fiducial marker for mapping the polypeptide chain, and its location is of interest also in relation to understanding the assembly properties of HBcAg and HBeAg. Our experimental strategy of peptide-based difference mapping was based on the considerations that (i) HBcAg assembly tolerates N-terminal extensions, and (ii) small domains or subdomains have been visualized in cryo-EM-derived density maps at relatively low resolutions (23–25).
MATERIALS AND METHODS
Capsids: Expression, Purification, and Assembly.
Plasmid ptacHpaII directs the synthesis of a 154-residue protein (Cpe), which includes the first ten residues of β-galactosidase, residues 3–144 of HBcAg, and two extraneous residues at the carboxy terminus (26). This plasmid was expressed in Escherichia coli, and the resulting protein was purified and assembled into capsids, as described (11, 16).
Cryo-EM.
Cpe capsid samples were frozen in thin films of vitrified ice suspended on holey carbon films (16). Micrographs were recorded at ×38,000 on a CM200-FEG (Philips, Eindhoven, the Netherlands) operating at 120 kV and fitted with a liquid-N2-cooled model 626 cryoholder and anticontamination blades (Gatan, Pleasanton, CA). Focal pairs of micrographs were recorded by using low-dose techniques. The focal pairs chosen for analysis had first contrast transfer function zeroes at spatial frequencies of (20 Å)−1 and (23 Å)−1 compared with (17 Å)−1 and (27 Å)−1 for the Cp147 data (2).
Image Reconstruction.
Micrographs of Cpe capsids were digitized on an SCAI scanner (Zeiss) at 7 μm per pixel to give a final sampling rate of 1.8 Å per pixel at the specimen. Individual particle images were extracted and processed by using a semiautomated procedure (27). The contrast transfer function was corrected and focal pairs were combined, as described (16, 2). Particle orientations were determined by means of the PFT (Polar Fourier Transform) algorithm (28) taking as starting models preexisting maps of Cp147 capsids (T = 4 (2); T = 3, unpublished results). The maps were calculated by using methodology reviewed in ref. 29. The numbers of particles included in each reconstruction were as follows: T = 4 (Cpe, n = 208; Cp147, n = 600); T = 3 (Cpe, n = 419; Cp147, n = 310). Resolutions were assessed in terms of a Fourier ring correlation method (2).
Difference maps were obtained as follows: a contour value was determined that enclosed 100% of the expected volume for the Cp147 capsid, given the particle mass and a partial specific volume of 0.78 Å3/Da (i.e., 3.8 × 106 Å3 for T = 3 and 5.0 × 106 Å3 for T = 4). Spherically averaged radial density profiles were calculated for both maps, and normalization of the Cpe map was adjusted to match the fit between these profiles. Difference maps were then obtained by subtraction. For surface rendering of the resulting maps, the following steps were taken: (i) inside a radius of 77 Å (T = 3) or 100Å (T = 4), the density was set to zero; (ii) small islands of density were filtered out, i.e., densities in them were set to zero. Surface views were created by using the visualization package of Advanced Visual Systems, Inc. (Waltham, MA), running on a Silicon Graphics (Mountain View, CA) workstation.
RESULTS
Our difference-imaging experiment compared Cpe and Cp147, two variants of the assembly domain. Their terminal sequences are given in Fig. 1. Disregarding which amino acids occupy positions 1, 2, 145, and 146, Cpe has eight more residues at its N terminus and one fewer residue at its C terminus. To a first approximation, therefore, the two proteins differ through the presence of an additional octapeptide at the N terminus of Cpe.
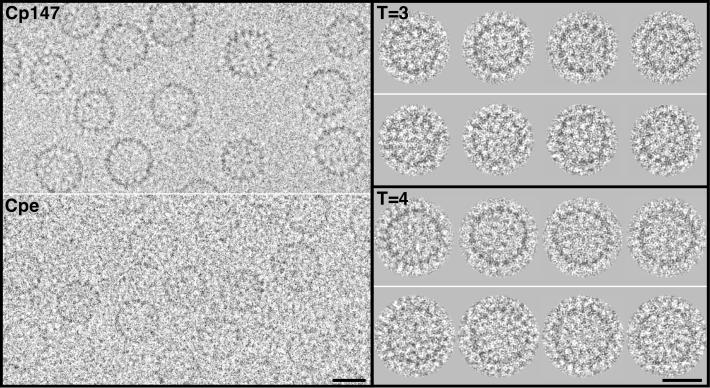
Cryoelectron micrographs of Cpe and Cp147 capsids are shown in Fig. 2. Both constructs produce both T = 3 (small) and T = 4 (large) capsids. Sixty-five percent of Cp147 capsids are T = 4, whereas only 15% of Cpe capsids are of this size (16). Nevertheless, each construct produced enough capsids of both sizes to allow two independent difference-imaging experiments, one for each size of capsid. No difference between Cpe and Cp147 capsids of a given size is evident from visual inspection (Fig. 2).
Figure 2.
(Left) Cryoelectron micrographs of Cp147 capsids and Cpe capsids. The capsids are of two sizes, with a higher incidence of larger (T = 4) capsids in the Cp147 preparation and of smaller (T = 3) capsids for Cpe. The Cpe micrograph was recorded closer to focus than the Cp147 micrograph, accounting for its lower contrast. (Upper Right) Gallery of four T = 3 capsids of Cp147 (upper row), compared with four Cpe capsids (lower row). (Lower Right) A similar comparison for T = 4 capsids. These images were obtained by computationally combining focal pairs and correcting for the contrast transfer function (Materials & Methods). As a result, the difference in contrast between the original micrographs (Left) was largely nullified. In this contrast transfer-function correction, the systematic attenuation of high frequencies (2) was not compensated, although this operation was performed in calculating the three-dimensional density maps. (Bars = 200 Å.)
To ensure an unbiased comparison, Cpe capsids were imaged under conditions similar to those previously used for Cp147, and the density maps were calculated under standardized conditions (2). We estimate the resolution of both reconstructions (T = 3 and T = 4) to be 11 Å. As reference structures, we used our T = 4 Cp147 map [(2); currently at a resolution of 7.9 Å; J.F.C., unpublished results], after bandlimiting it to the same resolution and a T = 3 Cp147 map, also at ≈11 Å resolution, which has not been published before. After imposing a common normalization, difference maps were calculated for both T = 4 and T = 3 capsids.
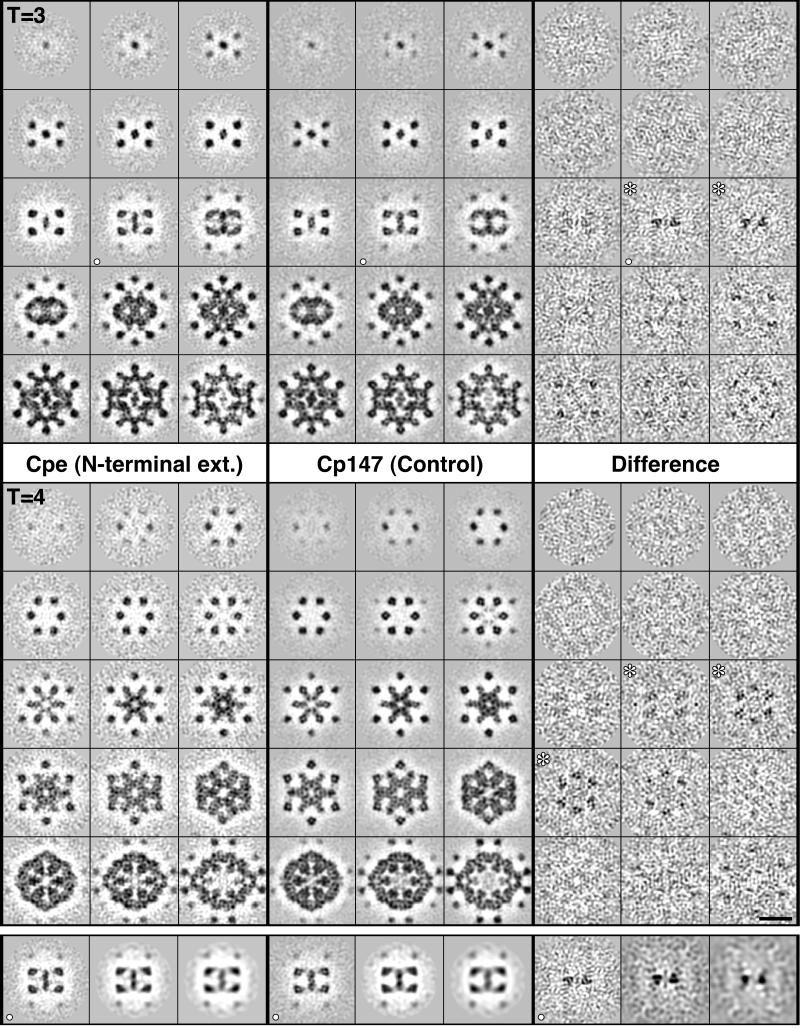
Serial sections through both difference maps are presented in Fig. 3. This analysis accommodates a 7-fold redundancy in the sense that the T = 3 and T = 4 capsids were calculated independently, and there are three quasiequivalent sites on the T = 3 capsid and four quasiequivalent sites on the T = 4 capsid. The density at each of these sites was calculated independently in the reconstructions, and the results obtained are mutually consistent. Significant positive density is seen in the difference maps only in the vicinity of the 3-fold axes and at a radius from the capsid center that corresponds to the outer surface of the contiguous shell (Fig. 3).
Figure 3.
Serial sections through density maps of Cpe and Cp147 capsids, as viewed along a 2-fold symmetry axis. The sections are spaced 3.7 Å apart. In each block of 3 × 5 panels, they run from the outer tip of a spike (upper left) through to the inner surface of the contiguous shell (lower right). Data are shown for both T = 3 (Top) and T = 4 (Middle) capsids. Protein (positive density) is dark. Shown at right are corresponding sections through the T = 3 and T = 4 difference maps. Significant density above background is seen in the sections marked with white asterisks. This density starts at radii −26 Å lower than the spike tips. In the T = 3 capsid, such density is seen at two local 3-fold axes and at both, it is 3-fold symmetric without imposition of this symmetry. The localized density seen in the fifth row of sections is also at (other) local 3-fold sites. In the T=4 difference map, positive densities are seen at the same distance below the spike tip: again, they are 3-fold symmetric (see Fig. 4), althugh in this case, the symmetry is less evident because the sectioning plane is less perpendicular to the local symmetry axes. In the bottom row of panels, section (○) through the T=3 capsid is shown as restricted in resolution from 11 Å to 15 Å and to 20 Å. Significant positive density persists in the difference map, although at 20 Å resolution, it is no longer resolved into three separate elements. (Bar = 100 Å.)
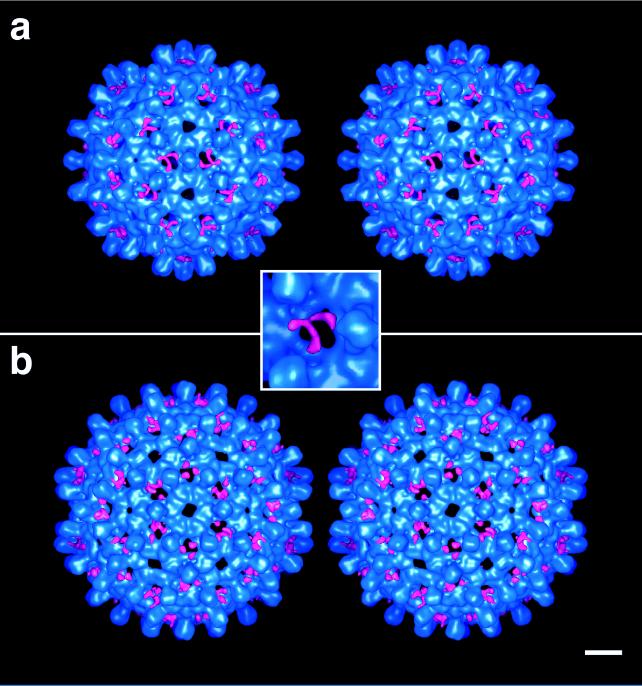
To convey the locations of this additional density, surface renderings of the capsids are presented in stereo in Fig. 4. In these representations, the reference Cp147 capsids are encoded in blue, and the additional density present on Cpe capsids is encoded in red. This density is clearly visible in the 60 holes at the local 3-fold axes of the T = 3 capsid and is absent from the holes on the 5-fold and global 3-fold axes of that capsid. Similarly, on the T = 4 capsid, this density is present at the holes on the local and global 3-fold axes and absent from the holes at the 5-fold and quasi-6-fold axes. In each case, three separate strands of density are seen, one emanating from each subunit around the hole in question. We conclude that these strands represent three copies of the extraneous octapeptide and note that they emerge from either side of the hammer-headed dimer near the point at which the spike protrusion enters the contiguous shell. A consensus site for the N terminus of the assembly domain, thus localized, is marked on a model of the dimer in Fig. 5.
Figure 4.
Stereo pairs showing the outer surfaces of Cpe capsids for T = 3 (a) and T = 4 (b). Coded in blue is the structure that is common to the Cpe and Cp147 capsids, whereas the additional density present on the Cpe capsids is red. (Bar = 50 Å.)
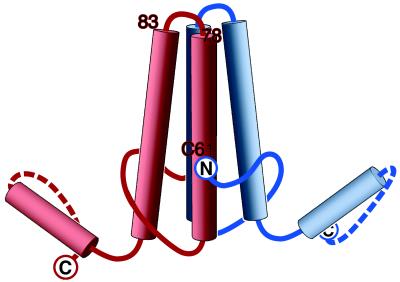
Figure 5.
Model of the HBV capsid protein dimer. The locations of the amino acids that have now been localized are marked: (i) the N terminus (this study), shown with the polypeptide chain at this point oriented as suggested by our difference maps (Fig. 4); (ii) the C terminus (5); and (iii) the loop covering residues 78–83 (7). The position of Cys 61 has also been inferred (Discussion). In most respects, this model confirms and supports that of Böttcher et al. (1), but has been revised in light of more recent information. The handedness, originally assigned arbitrarily (1), has been switched on the basis of an experimental determination (7). There are also minor differences in the positions assigned to residue 149, which we assign to the inner surface with the chain direction as shown, and the N terminus, which we place ≈10 Å farther up from the bottom of the molecule, as shown.
DISCUSSION
The approach that we have used to determine the site occupied by the N terminus of HBcAg appears to have considerable potential as a method for localizing specific residues on proteins of interest by inserting peptides of 10 to 20 residues at targeted sites and detecting them as bulk labels by cryo-EM. Moreover, this observation has implications for regulation of HBV assembly.
Residue Localization by Difference Imaging Based on Peptide Insertions or Deletions.
Genetic engineering of proteins readily allows residues to be inserted or deleted at designated sites. We now consider the size, sequence, and likely insertion sites of peptides for difference-imaging experiments. Termini, particularly C termini, are often surface exposed and relatively unconstrained and should make promising targets. Surface loops should also be promising insertion points, provided they are not located at inter-subunit interfaces. In this context, we note that extended hydrophilic sequences are likely to be surface exposed, as are sites known to be sensitive to proteolysis or other posttranslational modifications. However, in dealing with an unknown structure, it is not generally known a priori which amino acids reside in surface loops. In practice, a certain amount of trial and error is likely to be needed and limited proteolysis in conjunction with microsequencing should provide an effective way of probing for amenable insertion sites. Alternatively, candidates may be identified from multiple sequence alignments of related proteins. For example, this approach reveals the feasibility of insertions at position 84 in the loop at the tip of the HBcAg spike (see Fig. 1 of ref. 22) and will be applicable to increasing numbers of proteins as the sequence database continues to expand.
Two important parameters are the sizes of peptides for insertion or deletion and the resolution required to visualize them. The smaller the peptide, the more precise a localization may be anticipated. On the other hand, the detectability of smaller peptides will be more sensitive to noise. Previously, a 12-kDa subunit was detected by difference imaging of herpesvirus capsids at 30–35 Å resolution (24, 25) and a loop of ≈3 kDa was detected by comparing a cryo-EM map of adenovirus at 30 Å resolution with a 3 Å map of its hexons (23). Here we have been able to detect reproducibly an octapeptide of less than 1 kDa in maps at ≈11 Å resolution. Resolution of this order is not an absolute requirement to visualize an octapeptide, as the additional density remained clearly visible in our difference map when the resolution was limited to 15 Å or even 20 Å (see Fig. 3 Bottom).
How precise is our localization? Assigning the N terminus to the point at which the additional density seen in the difference map meets the surface of the unmodified capsid (Fig. 4), we estimate this point to be specified to within ±5 Å or so in all three dimensions. The difference peptide is ≈7 Å in apparent width (Fig. 4) so that its center should be within ≈4 Å of either edge. Although this estimate of precision is smaller than our resolution (11 Å), we note that localization depends on detecting the position of the center of mass of a feature seen in the difference map, which is less reliant on resolution per se.
The relationship between resolution and visibility of mobile elements on proteins poses an interesting paradox that invites further study. Although mobile elements are generally not visible in high-resolution crystallographic density maps, some of them should be visible in cryo-EM-derived maps at lower resolution (23), albeit not in full conformational detail. In practice, their visibility will depend on such factors as size, the dimensional scale of the disordering, and the resolution and statistical quality of the density map.
The amino acid sequence inserted may also be optimized for the purpose at hand. (In the present work, the construct used was readily available and no effort was made to optimize the appended sequence.) Ideally, the insert should assume a compact conformation, preferably a loop, so that the entry and exit points are in close proximity. Possibly, hydrophobic sequences may be suitable for this purpose because they should assume compact conformations, although they may compromise solubility. In principle, a peptide of ≈10–20 residues with a compact conformation and its two ends close together would be ideal; thus, the beta-hairpin (30) or a short loop stabilized by a disulfide bond appear to be promising candidates.
Finally, the imposed change in primary structure should not alter the fold or oligomerization state of the rest of the protein if a clearcut answer is to be obtained by difference imaging. After a construct has been obtained, useful indications that the desired fold has been achieved can be obtained by optical spectroscopy and for its oligomerization state by sedimentation analysis. Ultimately, a more stringent condition for isomorphous insertion is that the difference map obtained should contain only a single localized peak of additional density. This condition was met in the current analysis (Fig. 3).
Distinction Between HBcAg and HBeAg Antigens.
Substantial additions may be effected at the N terminus of the HBV capsid protein without impairing its ability to polymerize (19, 20). Nevertheless, native HBeAg, with its 10-residue N-terminal extension, remains unassembled in vivo, although other constructs with similarly sized extensions, such as Cpe, do assemble readily. It has been determined that Cys −7 in the residual propeptide forms an intramolecular disulfide bond with Cys 61 (31, 32), and it has been inferred that this bond somehow impedes assembly. In native HBcAg lacking Cys −7, Cys 61 forms an intermolecular disulfide bond with the same residue in the other subunit. This disulfide is not required for folding or assembly because proteins in which Cys 61 is substituted do assemble correctly (32), but presumably it reinforces an already stable capsid.
We have localized the N terminus to the side of the spike (Fig. 5). This assignment places it close to Cys 61 according to the following argument: residues 78–83 form the loop at the top of the spike (7) and the known location of the C terminus (5) indicates which of the two helices that emerge from the loop leads to the C terminus and, conversely, which leads to the N terminus. The latter helix is primarily responsible for intersubunit interactions at the dimer interface. Thus, Cys 61 should be ≈26 Å (17 residues at 1.54 Å per residue) along this helix from the spike tip, placing it just inside the contiguous shell and quite close to the N terminus of the same subunit (Fig. 5). In essence, the above arguments recapitulate those of Böttcher et al. (1) but are bolstered by experimental localization of N terminus and that inferred for Cys 61.
We do not yet know the azimuthal setting of Cys 61 around the helical axis, but presumably it is poised to interact across the dimer interface to form the Cys 61/Cys 61 disulfide bond (Fig. 5). In principle, a cysteine connected to the N terminus by a six-residue linker could interact with Cys 61 on the same subunit in one of two ways: (i) by reaching across the dimer interface; or (ii) by folding back around the outside of the spike. In option i, the linker would be likely to impede dimer formation and thus capsid assembly (1). However, this argument assumes that unpolymerized HBeAg is monomeric and such is not the case, as we have found (unpublished work) that the HBeAg construct (residues −10 to 149), expressed in E. coli, is a dimer at physiological ionic strength and neutral pH, under both reducing and oxidizing conditions. Moreover, the propensity of this dimer for assembly, although diminished compared with Cpe, is not eliminated. Thus, mechanism ii appears more likely.
Several possibilities remain open to explain the nonparticulate nature of native HBeAg. It may be that the protein is exported from the cytoplasm so rapidly that it never builds up a critical concentration for capsid assembly, or that the native propeptide causes the precursor protein to assume a conformation that prevents it from becoming assembly competent, or that conformational effects accompanying formation of the Cys 61/Cys −7 disulfide bond, somehow transmitted through the molecule, affect the polymerization sites at the tips of the dimer arms.
Acknowledgments
We thank Drs. B. L. Trus and D. Belnap for support and collegial discussions; Dr. T. S. Baker (Purdue University) for software; Dr. C. Hyde for making available a graphics workstation; and J. Kaufman and I. Palmer for preparing genetic constructs and purifying proteins.
ABBREVIATIONS
- cryo-EM
cryoelectron microscopy
- HBV
hepatitis B virus
References
- 1.Böttcher B, Wynne S A, Crowther R A. Nature (London) 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 2.Conway J F, Cheng N, Zlotnick A, Wingfield P T, Stahl S J, Steven A C. Nature (London) 1997;386:91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- 3.Trus B L, Roden R B S, Greenstone H L, Vrhel M, Schiller J T, Booy F P. Nat Struct Biol. 1997;4:413–420. doi: 10.1038/nsb0597-413. [DOI] [PubMed] [Google Scholar]
- 4.Hainfeld J F. Science. 1987;236:450–453. doi: 10.1126/science.3563522. [DOI] [PubMed] [Google Scholar]
- 5.Zlotnick A, Cheng N, Stahl S J, Conway J F, Steven A C, Wingfield P T. Proc Natl Acad Sci USA. 1997;94:9556–9561. doi: 10.1073/pnas.94.18.9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang G J, Porta C, Chen Z G, Baker T S, Johnson J E. Nature (London) 1992;355:275–278. doi: 10.1038/355275a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conway J F, Cheng N, Zlotnick A, Stahl S J, Wingfield P T, Belnap D M, Kanngiesser U, Noah M, Steven A C. J Mol Biol. 1998;279:1111–1121. doi: 10.1006/jmbi.1998.1845. [DOI] [PubMed] [Google Scholar]
- 8.Blumberg B S. Proc Natl Acad Sci USA. 1997;94:7121–7125. doi: 10.1073/pnas.94.14.7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollinger F B. In: Fields Virology. 3rd Ed. Fields B N, Knipe D M, Powley P M, editors. Philadelphia: Lippincott; 1996. pp. 2738–2808. [Google Scholar]
- 10.Bartenschlager R, Schaller H. EMBO J. 1992;11:3413–3420. doi: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wingfield P T, Stahl S J, Williams R W, Steven A C. Biochemistry. 1995;34:4919–4932. doi: 10.1021/bi00015a003. [DOI] [PubMed] [Google Scholar]
- 12.Miyanohara A, Imamura T, Araki M, Sugawara K, Ohtomo N, Matsubara K. J Virol. 1986;59:176–180. doi: 10.1128/jvi.59.1.176-180.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seifer M, Standring D N. J Virol. 1993;67:249–257. doi: 10.1128/jvi.67.1.249-257.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caspar D L D, Klug A. Cold Spring Harbor Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 15.Crowther R A, Kiselev N A, Böttcher B, Berriman J A, Borisova G P, Ose V, Pumpens P. Cell. 1994;77:943–950. doi: 10.1016/0092-8674(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 16.Zlotnick A, Cheng N, Conway J F, Booy F P, Steven A C, Stahl S J, Wingfield P T. Biochemistry. 1996;35:7412–7421. doi: 10.1021/bi9604800. [DOI] [PubMed] [Google Scholar]
- 17.Birnbaum F, Nassal M. J Virol. 1990;64:3319–3330. doi: 10.1128/jvi.64.7.3319-3330.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seifer M, Standring D N. Intervirology. 1995;38:47–62. doi: 10.1159/000150414. [DOI] [PubMed] [Google Scholar]
- 19.Schodel F, Moriarty A M, Peterson D L, Zheng J A, Hughes J L, Will H, Leturcq D J, McGee J S, Milich D R. J Virol. 1992;66:106–114. doi: 10.1128/jvi.66.1.106-114.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pumpens P, Borisova G P, Crowther R A, Grens E. Intervirology. 1995;38:63–74. doi: 10.1159/000150415. [DOI] [PubMed] [Google Scholar]
- 21.Zhou S, Standring D N. Proc Natl Acad Sci USA. 1992;89:10046–10050. doi: 10.1073/pnas.89.21.10046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bringas R. J Struct Biol. 1997;118:189–196. doi: 10.1006/jsbi.1997.3846. [DOI] [PubMed] [Google Scholar]
- 23.Stewart P L, Fuller SD, Burnett R M. EMBO J. 1993;12:2589–2599. doi: 10.1002/j.1460-2075.1993.tb05919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Booy F P, Trus B L, Newcomb W W, Brown J C, Conway J F, Steven A C. Proc Natl Acad Sci USA. 1994;91:5652–5656. doi: 10.1073/pnas.91.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Z H, Prasad B V V, Jakana J, Rixon F J, Chiu W. J Mol Biol. 1994;242:456–469. doi: 10.1006/jmbi.1994.1594. [DOI] [PubMed] [Google Scholar]
- 26.Stahl S J, Murray K. Proc Natl Acad Sci USA. 1989;86:6283–6287. doi: 10.1073/pnas.86.16.6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conway J F, Trus B L, Booy F P, Newcomb W W, Brown J C, Steven A C. J Struct Biol. 1993;111:222–233. doi: 10.1006/jsbi.1993.1052. [DOI] [PubMed] [Google Scholar]
- 28.Baker T S, Cheng R H. J Struct Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- 29.Fuller S D, Butcher S J, Cheng R H, Baker T S. J Struct Biol. 1996;116:48–55. doi: 10.1006/jsbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- 30.Blanco F, Ramirez-Alvarado M, Serrano L. Curr Opin Struct Biol. 1998;8:107–111. doi: 10.1016/s0959-440x(98)80017-1. [DOI] [PubMed] [Google Scholar]
- 31.Wasenauer G, Kock J, Schlicht H J. J Virol. 1992;66:5338–5346. doi: 10.1128/jvi.66.9.5338-5346.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nassal M, Rieger A. J Virol. 1993;67:4307–4315. doi: 10.1128/jvi.67.7.4307-4315.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]