Abstract
To create a universal system for the control of gene expression, we have studied methods for the construction of novel polydactyl zinc finger proteins that recognize extended DNA sequences. Elsewhere we have described the generation of zinc finger domains recognizing sequences of the 5′-GNN-3′ subset of a 64-member zinc finger alphabet. Here we report on the use of these domains as modular building blocks for the construction of polydactyl proteins specifically recognizing 9- or 18-bp sequences. A rapid PCR assembly method was developed that, together with this predefined set of zinc finger domains, provides ready access to 17 million novel proteins that bind the 5′-(GNN)6-3′ family of 18-bp DNA sites. To examine the efficacy of this strategy in gene control, the human erbB-2 gene was chosen as a model. A polydactyl protein specifically recognizing an 18-bp sequence in the 5′-untranslated region of this gene was converted into a transcriptional repressor by fusion with Krüppel-associated box (KRAB), ERD, or SID repressor domains. Transcriptional activators were generated by fusion with the herpes simplex VP16 activation domain or with a tetrameric repeat of VP16’s minimal activation domain, termed VP64. We demonstrate that both gene repression and activation can be achieved by targeting designed proteins to a single site within the transcribed region of a gene. We anticipate that gene-specific transcriptional regulators of the type described here will find diverse applications in gene therapy, functional genomics, and the generation of transgenic organisms.
Since Jacob and Monod questioned the chemical nature of the repressor and proposed a scheme by which the synthesis of individual proteins within a cell might be “provoked” or “repressed,” specific experimental control of gene expression has been a tantalizing prospect (1). It is now well established that genomes are regulated at the level of transcription primarily through the action of proteins known as transcription factors that bind DNA in a sequence-specific fashion. Often these protein factors act in a complex combinatorial manner allowing temporal, spatial, and environmentally responsive control of gene expression (2). Transcription factors frequently act both through a DNA-binding domain that localizes the protein to a specific site within the genome and through accessory effector domains that act to provoke (activate) or repress transcription at or near that site (3). Effector domains, such as the activation domain VP16 (4) and the repression domain Krüppel-associated box (KRAB) (5), are typically modular and retain their activity when they are fused to other DNA-binding proteins. Whereas genes might be readily controlled by directing transcription factors to particular sites within a genome, the design of DNA-binding proteins that might be fashioned to bind any given sequence has been a daunting challenge.
Our approach to this challenge is based on the recognition of the structural features unique to the Cys2-His2 class of nucleic acid-binding, zinc finger proteins. The Cys2-His2 zinc finger domain consists of a simple ββα fold approximately 30 amino acids in length. Structural stability of this fold is achieved by hydrophobic interactions and by chelation of a single zinc ion by the conserved Cys2-His2 residues (6). Nucleic acid recognition is achieved through specific amino acid side chain contacts originating from the α-helix of the domain, which typically binds 3 bp of DNA sequence (7, 8). Unlike other nucleic acid recognition motifs, simple covalent linkage of multiple zinc finger domains allows the recognition of extended asymmetric sequences of DNA. Studies of natural zinc finger proteins have shown that three zinc finger domains can bind 9 bp of contiguous DNA sequence (7, 9). Whereas recognition of 9 bp of sequence is insufficient to specify a unique site within even the small genome of Escherichia coli, we have demonstrated that polydactyl proteins containing six zinc finger domains can specify 18-bp recognition (10). With respect to the development of a universal system for gene control, an 18-bp address can be sufficient to specify a single site within all known genomes. Polydactyl proteins of this type are unknown in nature; however, we recently demonstrated their efficacy in gene activation and repression within living human cells (10).
Herein, we demonstrate the simplicity and efficacy of a general strategy for the rapid production of gene switches. With a family of defined zinc finger domains recognizing sequences of the 5′-GNN-3′ subset of a 64-member zinc finger alphabet (unpublished results), polydactyl proteins specifically recognizing novel 9- or 18-bp sequences were constructed and characterized. Potent transcription factors were generated and shown to control both gene activation and repression. Gene activation was achieved by using the herpes simplex virus VP16 activation domain (4) and a recombinant tetrameric repeat of its minimal activation domain. Gene repression or silencing was achieved by using three effector domains of human origin, the Krüppel-associated box (KRAB) (5), the ERF repressor domain (ERD) (12), and the mSIN3 interaction domain (SID) (13). Using luciferase reporter gene assays in human epithelial cells, we show that artificial transcriptional regulators, designed to target the promoter of the protooncogene erbB-2/HER-2, can ablate or activate gene expression in a specific manner. Gene activation or repression was achieved by targeting within the gene transcript, suggesting that information obtained from expressed sequence tags (ESTs) may be sufficient for the construction of gene switches. The methodology and materials described here promise diverse applications in gene therapy, transgenic organisms, functional genomics, and other areas of cell and molecular biology.
MATERIALS AND METHODS
Generation of Polydactyl Proteins with Desired DNA-Binding Specificity.
The studies reported here use the finger 2 (F2) variants pmGAC, pmGAG, pGCA, pGCC, pmGGA, pmGGC, pmGGG, and pGTG. To generate DNAs encoding three-finger proteins, F2 coding regions were PCR amplified from selected or designed F2 variants and assembled by PCR overlap extension. Alternatively, DNAs encoding three-finger proteins with a Zif268 or Sp1C framework were synthesized from 8 or 6 overlapping oligonucleotides, respectively. Sp1C framework constructs, used for all reporter assays described in this paper, were generated as follows. In the case of E2C-HS1(Sp1), 0.4 pmol each of oligonucleotides SPE2–3 (5′-GCG AGC AAG GTC GCG GCA GTC ACT AAA AGA TTT GCC GCA CTC TGG GCA TTT ATA CGG TTT TTC ACC-3′) and SPE2–4 (5′-GTG ACT GCC GCG ACC TTG CTC GCC ATC AAC GCA CTC ATA CTG GCG AGA AGC CAT ACA AAT GTC CAG AAT GTG GC-3′) were mixed with 40 pmol each of oligonucleotides SPE2–2 (5′-GGT AAG TCC TTC TCT CAG AGC TCT CAC CTG GTG CGC CAC CAG CGT ACC CAC ACG GGT GAA AAA CCG TAT AAA TGC CCA GAG-3′) and SPE2–5 (5′-ACG CAC CAG CTT GTC AGA GCG GCT GAA AGA CTT GCC ACA TTC TGG ACA TTT GTA TGG C-3′) in a standard PCR mixture and cycled 25 times (30 sec at 94°C, 30 sec at 60°C, 30 sec at 72°C). An aliquot of this preassembly reaction mixture was then amplified with 40 pmol each of the primers SPE2–1 (5′-GAG GAG GAG GAG GTG GCC CAG GCG GCC CTC GAG CCC GGG GAG AAG CCC TAT GCT TGT CCG GAA TGT GGT AAG TCC TTC TCT CAG AGC-3′) and SPE2–6 (5′-GAG GAG GAG GAG CTG GCC GGC CTG GCC ACT AGT TTT TTT ACC GGT GTG AGT ACG TTG GTG ACG CAC CAG CTT GTC AGA GCG-3′), using the same cycling conditions. The E2C-HS2(Sp1) DNA was generated in the same way, using an analogous set of oligonucleotides differing only in the recognition helix coding regions. All assembled three-finger coding regions were digested with the restriction endonuclease SfiI and cloned in pMal-CSS, a derivative of the bacterial expression vector pMal-C2 (New England Biolabs). DNAs encoding six-finger proteins with each of the different frameworks were assembled in pMal-CSS by using XmaI and BsrF1 restriction sites included in the sequences flanking the three-finger coding regions (for the positions of the restriction sites, see Fig. 3). Each of the zinc finger proteins was expressed in the E. coli strain XL1-Blue, and binding properties were investigated by ELISA and gel-shift analysis.
Figure 3.
Amino acid sequence alignment of six-finger proteins specific for the 18-bp target sequence e2c. Recognition helix positions −2 to 6 of each finger (F1 to F6) are underlain with dark gray boxes and labeled according to their DNA-binding specificity. Sequence identities in the framework regions are underlain with light gray boxes. Positions of the conserved cysteine and histidine residues are marked by asterisks. The corresponding positions of the SfiI recognition sites, used for cloning the zinc finger coding regions into the various expression vectors, as well as the BsrF1 and XmaI recognition sites, used for the construction of DNAs encoding six-finger proteins, are indicated.
Construction of Zinc Finger–Effector Domain Fusion Proteins.
For the construction of zinc finger–effector domain fusion proteins, DNAs encoding amino acids 473–530 of the ets2 repressor factor (ERF) repressor domain (ERD) (12), amino acids 1–97 of the KRAB domain of KOX1 (5), or amino acids 1–36 of the Mad mSIN3 interaction domain (SID) (13) were assembled from overlapping oligonucleotides by using Taq DNA polymerase. The coding region for amino acids 413–489 of the VP16 transcriptional activation domain (4) was PCR amplified from pcDNA3/C7-C7-VP16 (10). The VP64 DNA, encoding a tetrameric repeat of VP16’s minimal activation domain, comprising amino acids 437–447 (14), was generated from two pairs of complementary oligonucleotides. The resulting fragments were fused to zinc finger coding regions by standard cloning procedures, such that each resulting construct contained an internal simian virus 40 nuclear localization signal, as well as a C-terminal hemagglutinin (HA) decapeptide tag. Fusion constructs were cloned in the eukaryotic expression vector pcDNA3 (Invitrogen).
Construction of Luciferase Reporter Plasmids.
An erbB-2 promoter fragment comprising nucleotides −758 to −1, relative to the ATG initiation codon, was PCR amplified from human bone marrow genomic DNA with the TaqExpand DNA polymerase mix (Boehringer Mannheim) and inserted into pGL3basic (Promega), upstream of the firefly luciferase gene. A human erbB-2 promoter fragment encompassing nucleotides −1571 to −24 was excised from pSVOALΔ5′/erbB-2(N-N) (15) by HindIII digestion and subcloned into pGL3basic, upstream of the firefly luciferase gene.
Luciferase Assays.
For all transfections, HeLa cells were used at a confluency of 40–60%. Typically, cells were transfected with 400 ng of reporter plasmid (pGL3-promoter constructs or, as negative control, pGL3basic), 50 ng of effector plasmid (zinc finger constructs in pcDNA3 or, as negative control, empty pcDNA3), and 200 ng of internal standard plasmid (phrAct-βGal) in a well of a six-well dish by using the Lipofectamine reagent (GIBCO/BRL). Cell extracts were prepared approximately 48 hr after transfection. Luciferase activity was measured with luciferase assay reagent (Promega), β-galactosidase activity with Galacto-Light (Tropix), in a MicroLumat LB96P luminometer (EG & G Berthold, Gaithersburg, MD). Luciferase activity was normalized to β-galactosidase activity.
RESULTS AND DISCUSSION
The erbB-2 Gene as a Target for Zinc Finger-Based Transcriptional Control.
The human erbB-2 gene was chosen as a model target for the development of zinc finger-based transcriptional switches. Members of the ErbB receptor family play important roles in the development of human malignancies. In particular, erbB-2 is overexpressed as a result of gene amplification and/or transcriptional deregulation in a high percentage of human adenocarcinomas arising at numerous sites, including breast, ovary, lung, stomach, and salivary gland (16). Increased expression of ErbB-2 leads to constitutive activation of its intrinsic tyrosine kinase and has been shown to cause the transformation of cultured cells. Numerous clinical studies have shown that patients bearing tumors with elevated ErbB-2 expression levels have a poorer prognosis (16). In addition to its involvement in human cancer, erbB-2 plays important biological roles, both in the adult and during embryonal development of mammals (16–18).
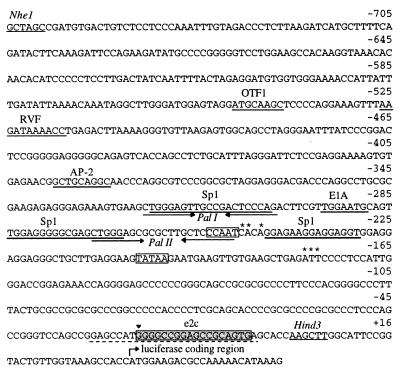
The erbB-2 promoter therefore represents an interesting test case for the development of artificial transcriptional regulators. This promoter has been characterized in detail and has been shown to be relatively complex, containing both a TATA-dependent and a TATA-independent transcriptional initiation site (19). Whereas our early studies showed that polydactyl proteins could act as transcriptional regulators that specifically activate or repress transcription, these proteins bound upstream of an artificial promoter to six tandem repeats of the proteins’ binding site (10). Furthermore, this study utilized polydactyl proteins that were not modified in their binding specificity. Herein, we wished to test the efficacy of polydactyl proteins assembled, from predefined building blocks, to bind a single site in the native erbB-2 promoter. We have generated and characterized a family of zinc finger domains that bind each of the 16 5′-GNN-3′ DNA triplets. One reason we focused on the production of this family of recognition domains is that promoter regions of most organisms are relatively G+C rich. Thus, if proteins recognizing 5′-(GNN)x-3′ sites could be readily assembled from this set of defined zinc finger domains, many genes could be rapidly and specifically targeted for regulation. A protein containing six zinc finger domains and recognizing 18 bp of DNA should be sufficient to define a single address within all known genomes. Examination of the erbB-2 promoter region revealed two 5′-(GNN)6-3′ sites and one 5′-(GNN)9-3′ site. One of these sites, identified here as e2c, falls within the 5′ untranslated region of the erbB-2 gene and was chosen as the target site for the generation of a gene-specific transcriptional switch (Fig. 1). A blast sequence similarity search of the GenBank database confirmed that this sequence is unique to erbB-2. The position of the e2c target sequence, downstream and in the vicinity of the two major transcription initiation sites, would allow us to examine repression through inhibition of either transcription initiation or elongation. Further, if we could demonstrate the ability to modulate gene expression by targeting within the transcribed region of a gene, EST data might be used in the design of transcriptional regulators, obviating the need to sequence the promoter region to regulate the gene. An interesting feature of the e2c target site is that it is found within a short stretch of sequence that is conserved between human, rat, and mouse erbB-2 genes (20). Thus, targeting of this site would allow for the study of this strategy in animal models prior to its application to human disease.
Figure 1.
Nucleotide sequence of the human erbB-2 promoter fragment used in these studies. Nucleotide positions −758 to −1 relative to the ATG initiation codon are shown, with known transcription factor binding sites (16) underlined. Sp1 binding sites are marked as mapped by DNase footprinting (32). The two palindromic sequences Pal I and II bound by RBPJκ (11) are marked by inverted arrows. CCAAT and TATAA sequences are boxed. The major transcription initiation sites (19) are indicated by asterisks. The zinc finger target sequence e2c is underlain with a gray box. The short stretch of sequence identity between human, rat, and mouse genes is indicated by a dashed underline. Restriction sites used for cloning of the promoter fragment are indicated. The arrowhead at position −24 denotes the 3′ end of an erbB-2 control promoter fragment lacking the zinc finger target sequence e2c.
Modular Strategies for the Generation of Polydactyl Proteins.
For generating polydactyl proteins with desired DNA-binding specificity we have focused on the assembly of predefined zinc finger domains, an approach that contrasts with the sequential selection strategy proposed by Greisman and Pabo (21). Such a strategy would require the sequential generation and selection of six zinc finger libraries for each required protein, making this experimental approach inaccessible to most laboratories and extremely time consuming to all. Further, because it is difficult to apply specific negative selection against binding alternative sequences in this strategy, proteins may result that are relatively unspecific, as was recently reported (22).
We investigated the general utility of two different strategies for generating three-finger proteins recognizing 9 bp of DNA sequence. Each strategy is based on the modular nature of the zinc finger domain and takes advantage of a family of zinc finger domains recognizing triplets of the 5′-GNN-3′ type. Two three-finger proteins recognizing half-sites (HS) 1 and 2 of the 5′-(GNN)6-3′ erbB-2 target site e2c (Fig. 1) were generated in the first strategy by fusing the predefined finger 2 (F2) domain variants together by using a PCR assembly strategy. To examine the generality of this approach, three additional three-finger proteins recognizing sequences of the 5′-(GNN)3-3′ type were prepared by the same approach. Purified zinc finger proteins were prepared as fusions with the maltose-binding protein (MBP). ELISA analysis revealed that serially connected F2 proteins were able to act in concert to specifically recognize the desired 9-bp DNA target sequences (Fig. 2A). Each of the five proteins shown was able to discriminate between target and nontarget 5′-(GNN)3-3′ sequence.
Figure 2.
ELISA analysis of zinc finger DNA-binding specificities. The indicated three-finger proteins (A) and six-finger proteins (B) were expressed in E. coli as MBP fusion proteins. Specificity of binding was analyzed by measuring the binding activity in total lysates to immobilized biotinylated hairpin oligonucleotides containing the indicated 9-bp (A) or 18-bp (B) targets. The nucleotide sequences of the six-finger nontarget oligonucleotides were as follows: e1a, 5′-GCC GAG GCG GCC GGA GTC-3′; e1b, 5′-GTT GTG GCG TTG GCG GCG-3′; b3, 5′-GCC TGA GAG GGA GCG GTG-3′; c5, 5′-GCG GAG GCA GGA GGC GGG-3′; zif-zif, 5′-GCG TGG GCG GCG TGG GCG-3′ (B). Assays were performed in duplicate and the maximal signals were normalized to 1. The open box on top of each bar represents the standard deviation.
The affinity of each of the proteins for its target was determined by electrophoretic mobility-shift assays. These studies demonstrated that the zinc finger peptides have affinities comparable to Zif268 and other natural transcription factors with Kd values that ranged from 3 to 70 nM (Table 1). Here we determined the Kd of Zif268 for its operator to be 10 nM. It must be noted that, for reasons that remain to be explained, one group has reported Kd values for the natural Zif268 protein that range from 6 nM to 10 pM, a 600-fold variation (7, 21). Most studies have reported the Kd of the Zif268–DNA interaction to be from 3 to 10 nM (23, 24). Thus, to compare the results reported here with those reported elsewhere, the relative Kd values should be compared, (mutant Kd)/(Zif268 Kd), where both values are derived from the same report. Our results compare favorably to other studies of novel three-finger proteins prepared by using phage display, where affinities 10- to 200-fold weaker than Zif268 were reported (21, 25).
Table 1.
Summary of three- and six-finger protein affinities
| Protein | Target | Target sequences (5′-3′) | Kd, nM |
|---|---|---|---|
| B3 (F2) | b3 | GGA GGG GAC g | 4 |
| E2 (F2) | e2 | GGG GGC GAG g | 3 |
| C5 (F2) | c5 | GGA GGC GGG g | 30 |
| E2C-HS1 (F2) | e2c-hs1 | GGG GCC GGA g | 45 |
| E2C-HS1 (Zif) | e2c-hs1 | GGG GCC GGA g | 70 |
| E2C-HS1 (Sp1) | e2c-hs1 | GGG GCC GGA g | 35 |
| E2C-HS2 (F2) | e2c-hs2 | GCC GCA GTG g | 70 |
| E2C-HS2 (Zif) | e2c-hs2 | GCC GCA GTG g | 75 |
| E2C-HS2 (Sp1) | e2c-hs2 | GCC GCA GTG g | 25 |
| E2C (F2) | e2c-g | GGG GCC GGA GCC GCA GTG g | 25 |
| E2C (Zif) | e2c-g | GGG GCC GGA GCC GCA GTG g | 1.6 |
| E2C (Zif) | e2c-a | GGG GCC GGA GCC GCA GTG a | 2.3 |
| E2C (Zif) | e2c-muths1 | AGT CTG AAT GCC GCA GTG g | 200 |
| E2C (Zif) | e2c-muths2 | GGG GCC GGA AGT CTG AAT g | 200 |
| E2C (Sp1) | e2c-g | GGG GCC GGA GCC GCA GTG G | 0.5 |
| E2C (Sp1) | e2c-a | GGG GCC GGA GCC GCA GTG a | 0.75 |
| E2C (Sp1) | e2c-muths1 | AGT CTG AAT GCC GCA GTG g | 65 |
| E2C (Sp1) | e2c-muths2 | GGG GCC GGA AGT CTG AAT g | 100 |
Affinities of three- and six-finger proteins for various target sequences were determined by gel shift analysis. Proteins are named with uppercase letters, DNA target sequences with lowercase letters. F2, finger 2 framework; Zif, Zif268 framework; Sp1, Sp1C framework; mut, mutant; Hs, half-site. With respect to the target site overlap phenomenon, the base following each target sequence is given as a lowercase letter. The affinity of the Zif268-DNA interaction was determined to be 10 nM (unpublished results). Kd values are averaged from two independent experiments, with standard deviations of 50% or less.
As an alternative to the serial connection of F2 domain variants, in the second strategy, three-finger proteins specific for the two e2c 5′-(GNN)3-3′ half-sites were produced by “helix grafting.” As shown in Fig. 3, the framework residues of the zinc finger domains, those residues that support the presentation of the recognition helix, vary between proteins. We anticipated that the framework residues may play a role in affinity and specificity. For helix grafting, amino acid positions −2 to 6 of the DNA recognition helices were grafted into either a Zif268 (7) or an Sp1C framework (26). The Sp1C protein is a designed consensus protein shown to have enhanced stability toward chelating agents. The proteins were expressed from DNA templates prepared by a rapid PCR-based gene assembly strategy. In each case, ELISA analysis of MBP fusion proteins showed that the DNA-binding specificities (Fig. 2A) and affinities observed with the F2 framework constructs were retained (Table 1).
Generation of Six-Finger Proteins for Specific Targeting of the erbB-2 Promoter Region.
As discussed above, the recognition of 9 bp of DNA sequence is not sufficient to specify a unique site within a complex genome. In contrast, a six-finger protein recognizing 18 bp of contiguous DNA sequence could define a single site in the human genome, thus fulfilling an important prerequisite for the generation of a gene-specific transcriptional switch. Six-finger proteins binding the erbB-2 target sequence e2c were generated from three-finger constructs by simple restriction enzyme digestion and cloning with F2, Zif268, and Sp1C framework template DNAs (Fig. 3). ELISA analysis of purified MBP fusion proteins showed that each of the six-finger proteins was able to recognize the specific target sequence, with little cross-reactivity to nontarget 5′-(GNN)6-3′ sites or a tandem repeat of the Zif268 target site (Fig. 2B).
The affinity of each protein for the e2c DNA target site was determined by gel-shift analysis. A modest Kd value of 25 nM was observed with the E2C(F2) six-finger protein constructed from the F2 framework (Table 1), a value that is only 2 to 3 times better than its constituent three-finger proteins. In our previous studies of six-finger proteins, we observed approximately 70-fold-enhanced affinity of the six-finger proteins for their DNA ligand as compared with their three-finger constituents (10). The absence of a substantial increase in the affinity of the E2C(F2) peptide suggested that serial connection of F2 domains is not optimal. It is possible that the periodicity of the F2 domains of the six-finger protein does not match that of the DNA over this extended sequence, and that a significant fraction of the binding energy of this protein is spent in unwinding DNA (27). In contrast to the F2 domain protein, the E2C(Zif) and E2C(Sp1) six-finger proteins displayed 40- to 70-fold-increased affinity as compared with their original three-finger protein constituents, with Kd values of 1.6 nM and 0.5 nM, respectively. Significantly, both three-finger components of these proteins were involved in binding, since mutation of either half-site led to a roughly 100-fold decrease in affinity (Table 1). The preponderance of known transcription factors bind their specific DNA ligands with nanomolar affinity, suggesting that the control of gene expression is governed by protein/DNA complexes of unexceptional lifetimes. Thus, zinc finger proteins of increased affinity should not be required and could be disadvantageous, especially if binding to nonspecific DNA is also increased.
The zinc finger domain is generally considered to be modular in nature, with each finger recognizing a 3-bp subsite (7). This idea is supported by our ability to recombine zinc finger domains in any desired sequence, yielding polydactyl proteins recognizing extended sequences of the structure 5′-(GNN)x-3′. However, it should be noted that, at least in some cases, zinc finger domains appear to specify overlapping 4-bp sites rather than individual 3-bp sites. In Zif268, residues in addition to those found at helix positions −1, 3, and 6 are involved in contacting DNA (8). Specifically, an aspartate in helix position 2 of F2 plays several roles in recognition and makes a variety of contacts. The carboxylate of the aspartate side chain hydrogen bonds with arginine at position −1, stabilizing its interaction with the 3′-guanine of its target site. This aspartate also participates in water-mediated contacts with the guanine’s complementary cytosine. In addition, this carboxylate is observed to make a direct contact to the N4 of the cytosine base on the opposite strand of the 5′-guanine base of the finger 1 binding site. It is this interaction that is the chemical basis for what we describe as target site overlap. Indeed, when the Zif268 F2 libraries were selected against the four 5′-GCG GNG GCG-3′ sequences, both an arginine at position −1 and an aspartate at position 2 were obtained, analogous to the residues in native Zif268. Since the e2c target sequence (5′-GGG GCC GGA GCC GCA GTG-3′) is followed by an A rather than a G, a potential target site overlap problem was anticipated with finger 1 of an e2c-specific six-finger protein. However, in both the Zif- and Sp1C-framework six-finger proteins, the GTG-specific finger 1 containing an aspartate at position 2 appears to recognize the sequences 5′-GTGA-3′ and 5′-GTGG-3′ equally well, as indicated by their very similar affinities to target sites e2c-a and e2c-g (Table 1).
Zinc Finger–Effector Domain Fusion Constructs for Specific Regulation of the erbB-2 Promoter.
To test the concept of using zinc finger proteins as gene-specific transcriptional regulators, the E2C(Sp1) six-finger protein was fused to a number of effector domains (Fig. 4). Transcriptional repressors were generated by attaching one of three human-derived repressor domains to the zinc finger protein. The first repressor protein was prepared with ERD (12), defined by amino acids 473–530 of the ets2 repressor factor (ERF). This domain mediates the antagonistic effect of ERF on the activity of transcription factors of the ets family. A synthetic repressor was constructed by fusion of this domain to the C terminus of the zinc finger protein. The second repressor protein was prepared with the KRAB domain (5). This repressor domain is commonly found at the N terminus of zinc finger proteins and presumably exerts its repressive activity on TATA-dependent transcription in a distance- and orientation-independent manner (28), by interacting with the RING finger protein KAP-1 (29). We utilized the KRAB domain found between amino acids 1 and 97 of the zinc finger protein KOX1 (5). In this case an N-terminal fusion with the six-finger protein was constructed. Finally, to explore the utility of histone deacetylation for repression, amino acids 1–36 of the Mad SID were fused to the N terminus of the zinc finger protein (13). This small domain is found at the N terminus of the transcription factor Mad and is responsible for mediating its transcriptional repression by interacting with mSIN3, which in turn interacts the corepressor N-CoR and with the histone deacetylase mRPD1 (30). To examine gene-specific activation, transcriptional activators were generated by fusing the zinc finger protein to amino acids 413–489 of the herpes simplex virus VP16 protein (4), or to an artificial tetrameric repeat of VP16’s minimal activation domain, DALDDFDLDML (14), termed VP64.
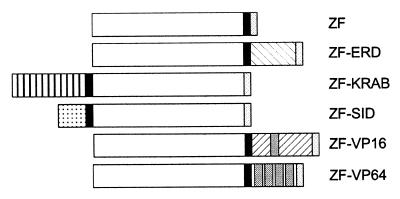
Figure 4.
Structure of zinc finger–effector domain fusion proteins. Effector domains are as labeled. White boxes, zinc fingers (ZF); black boxes, simian virus 40 nuclear localization signal; light gray boxes, hemagglutinin epitope tag; dark gray boxes, VP16 minimal activation domain.
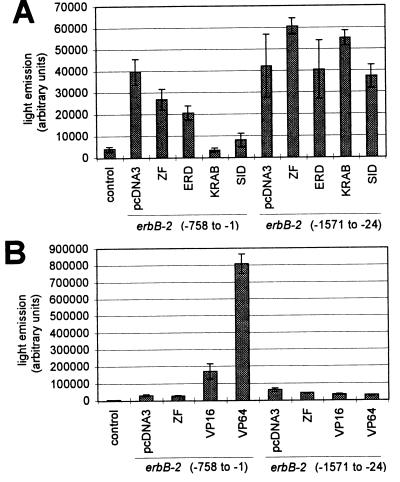
Reporter constructs containing fragments of the erbB-2 promoter coupled to a luciferase reporter gene were generated to test the specific activities of our designed transcriptional regulators. The target reporter plasmid contained nucleotides −758 to −1 with respect to the ATG initiation codon, whereas the control reporter plasmid contained nucleotides −1571 to −24, thus lacking all but one nucleotide of the E2C binding site encompassed in positions −24 to −7. Both promoter fragments displayed similar activities when transfected transiently into HeLa cells, in agreement with previous observations (15). To test the effect of zinc finger–repressor domain fusion constructs on erbB-2 promoter activity, HeLa cells were transiently cotransfected with each of the zinc finger expression vectors and the luciferase reporter constructs (Fig. 5A). Significant repression was observed with each construct. The ERD and SID fusion proteins produced approximately 50% and 80% repression, respectively. The most potent repressor was the KRAB fusion protein. This protein caused complete repression of erbB-2 promoter activity. The observed residual activity was at the background level of the promoterless pGL3 reporter. In contrast, none of the proteins caused significant repression of the control erbB-2 reporter construct lacking the E2C target site, demonstrating that repression is indeed mediated by specific binding of the E2C(Sp1) protein to its target site. Expression of a zinc finger protein lacking any effector domain resulted in weak repression, approximately 30%, indicating that most of the repression observed with the SID and KRAB constructs is caused by their effector domains, rather than by DNA binding alone. This observation strongly suggests that the mechanism of repression is active inhibition of transcription initiation rather than of elongation. Once initiation of transcription by RNA polymerase II has occurred, the zinc finger protein appears to be readily displaced from the DNA by the action of the polymerase.
Figure 5.
Specific repression (A) or activation (B) of erbB-2 promoter activity by using zinc finger–effector domain fusion proteins. HeLa cells were cotransfected with the indicated zinc finger expression plasmids and erbB-2 promoter–luciferase reporter constructs. The erbB-2 (−1571 to −24) reporter plasmid lacks the zinc finger target sequence. Luciferase activity in total cell extracts was measured 48 h after transfection. Each bar represents the mean value (±SD) of duplicate (A) or triplicate (B) measurements.
The utility of gene-specific polydactyl proteins to mediate activation of transcription was investigated with the same two reporter constructs (Fig. 5B). The VP16 fusion protein was found to stimulate transcription approximately 5-fold, whereas the VP64 fusion protein produced a 27-fold activation. This dramatic stimulation of promoter activity caused by a single VP16-based transcriptional activator is exceptional in view of the fact that the zinc finger protein binds in the transcribed region of the gene. This again demonstrates that mere binding of a zinc finger protein, even with one with subnanomolar affinity, in the path of RNA polymerase II need not necessarily negatively affect gene expression.
Conclusions.
We have demonstrated that zinc finger proteins capable of binding novel 9- and 18-bp DNA target sites can be rapidly prepared by using predefined domains recognizing 5′-GNN-3′ sites. While we have characterized only one-quarter of the 64 domains required for the recognition of any sequence, this information is sufficient for the preparation of 166 or 17 million novel six-finger proteins, each capable of binding 18 bp of DNA sequence. This rapid method for the construction of novel zinc finger proteins has advantages over the sequential generation and selection of zinc finger domains proposed by others (21) and takes advantage of structural information suggesting that the potential for the target overlap problem as defined above might be avoided in proteins targeting 5′-GNN-3′ sites. Using the complex and well studied erbB-2 promoter and live human cells, we have demonstrated that these proteins, when provided with the appropriate effector domain, can be used to “provoke” or activate expression and to produce graded levels of repression down to the level of the background in these experiments. Our studies suggest that the KRAB domain is significantly more potent as a transcriptional repressor than ERD or SID, and that it is able to inhibit both the TATA-dependent and the TATA-independent transcriptional initiation of this promoter. These repressor domains have not previously been directly compared. We believe that our strategy of using predefined zinc finger domains to construct polydactyl proteins coupled to effector domains has significant advantages over strategies that attempt to repress transcription only by competing or interfering with proteins involved in the transcription complex (22, 31). Utilization of effector domains that have the potential to act over a distance should allow the application of these gene switches to the regulation of uncharacterized genes and promoters. Because these transcriptional regulators might be prepared by using our PCR-assembly strategy in a high-throughput fashion, we believe it is appropriate to comment on their potential practical applications. Novel DNA-binding proteins generated in this manner should have potential utility in DNA-based diagnostic applications. For the study of gene function, we believe that the ability to both activate and repress the transcription of genes, at graded levels if necessary, may assist in assigning gene function. Since these proteins exert their control by acting in trans, functional gene knockout or activation might be produced in heterozygous transgenic animals. This would drastically reduce the time required to produce a gene knockout in a whole animal and would extend the range of organisms to which knockout technology might be applied. These proteins might also be used in gene therapy applications to inhibit the production of viral gene products or to activate genes involved in fighting disease. Significantly, the ease with which these proteins can be prepared will facilitate the testing of these ideas by the scientific community.
Acknowledgments
We thank J. Saldana, K. Bower, and M. Elia for their technical assistance, G. N. Gill for providing us with plasmids, and H. G. Bujard for discussions concerning the VP64 domain. This study was supported in part by National Institutes of Health grants to C.F.B., and R.R.B. was the recipient of postdoctoral fellowships from the Swiss National Science Foundation and the Krebsliga beider Basel.
ABBREVIATIONS
- ERD
ERF repressor domain
- F2
finger 2
- KRAB
Krüppel-associated box
- MBP
maltose-binding protein
- SID
mSIN3 interaction domain
References
- 1.Jacob F, Monod J. J Mol Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 2.Ptashne M. Nat Med. 1997;3:1069–1072. doi: 10.1038/nm1097-1069. [DOI] [PubMed] [Google Scholar]
- 3.Cowell I G. Trends Biochem Sci. 1994;19:38–42. doi: 10.1016/0968-0004(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 4.Sadowski I, Ma J, Triezenberg S, Ptashne M. Nature (London) 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 5.Margolin J F, Friedman J R, Meyer W K-H, Vissing H, Thiesen H-J, Rauscher F J., III Proc Natl Acad Sci USA. 1994;91:4509–4513. doi: 10.1073/pnas.91.10.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee M S, Gippert G P, Soman K V, Case D A, Wright P E. Science. 1989;245:635–637. doi: 10.1126/science.2503871. [DOI] [PubMed] [Google Scholar]
- 7.Pavletich N P, Pabo C O. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 8.Elrod-Erickson M, Rould M A, Nekludova L, Pabo C O. Structure. 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- 9.Swirnoff A H, Milbrandt J. Mol Cell Biol. 1995;15:2275–2287. doi: 10.1128/mcb.15.4.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Q, Segal D J, Ghiara J B, Barbas C F., III Proc Natl Acad Sci USA. 1997;94:5525–5530. doi: 10.1073/pnas.94.11.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Fischer W H, Gill G N. J Biol Chem. 1997;272:14110–14114. doi: 10.1074/jbc.272.22.14110. [DOI] [PubMed] [Google Scholar]
- 12.Sgouras D N, Athanasiou M A, Beal G J, Jr, Fisher R J, Blair D G, Mavrothalassitis G J. EMBO J. 1995;14:4781–4793. doi: 10.1002/j.1460-2075.1995.tb00160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ayer D E, Laherty C D, Lawrence Q A, Armstrong A P, Eisenman R N. Mol Cell Biol. 1996;16:5772–5781. doi: 10.1128/mcb.16.10.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seipel K, Georgiev O, Schaffner W. EMBO J. 1992;11:4961–4968. doi: 10.1002/j.1460-2075.1992.tb05603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hudson L G, Ertl A P, Gill G N. J Biol Chem. 1990;265:4389–4393. [PubMed] [Google Scholar]
- 16.Hynes N E, Stern D F. Biochim Biophys Acta. 1994;1198:165–184. doi: 10.1016/0304-419x(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 17.Altiok N, Bessereau J-L, Changeux J-P. EMBO J. 1995;14:4258–4266. doi: 10.1002/j.1460-2075.1995.tb00100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee K-F, Simon H, Chen H, Bates B, Hung M-C, Hauser C. Nature (London) 1995;378:394–398. doi: 10.1038/378394a0. [DOI] [PubMed] [Google Scholar]
- 19.Ishii S, Imamoto F, Yamanashi Y, Toyoshima K, Yamamoto T. Proc Natl Acad Sci USA. 1987;84:4374–4378. doi: 10.1073/pnas.84.13.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White M R-A, Hung M-C. Oncogene. 1992;7:677–683. [PubMed] [Google Scholar]
- 21.Greisman H A, Pabo C O. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- 22.Kim J-S, Pabo C O. J Biol Chem. 1997;272:29795–29800. doi: 10.1074/jbc.272.47.29795. [DOI] [PubMed] [Google Scholar]
- 23.Choo Y, Klug A. Proc Natl Acad Sci USA. 1994;91:11163–11167. doi: 10.1073/pnas.91.23.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamilton T B, Borel F, Romaniuk P J. Biochemistry. 1998;37:2051–2058. doi: 10.1021/bi9717993. [DOI] [PubMed] [Google Scholar]
- 25.Choo Y, Sanchez-Garcia I, Klug A. Nature (London) 1994;372:642–645. doi: 10.1038/372642a0. [DOI] [PubMed] [Google Scholar]
- 26.Desjarlais J R, Berg J M. Proc Natl Acad Sci USA. 1993;90:2256–2260. doi: 10.1073/pnas.90.6.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi Y, Berg J M. Biochemistry. 1996;35:3845–3848. doi: 10.1021/bi952384p. [DOI] [PubMed] [Google Scholar]
- 28.Pengue G, Lania L. Proc Natl Acad Sci USA. 1996;93:1015–1020. doi: 10.1073/pnas.93.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedman J R, Fredericks W J, Jensen D E, Speicher D W, Huang X-P, Neilson E G, Rauscher F J., III Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 30.Heinzel T, Lavinsky R M, Mullen T-M, Söderström M, Laherty C D, Torchia J, Yang W-M, Brard G, Ngo S D, Davie J R, et al. Nature (London) 1997;387:43–46. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 31.Kim J-S, Kim J, Cepek K L, Sharp P A, Pabo C O. Proc Natl Acad Sci USA. 1997;94:3616–3620. doi: 10.1073/pnas.94.8.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, Gill G N. Oncogene. 1994;9:2269–2276. [PubMed] [Google Scholar]