Abstract
Single-stranded DNA-binding proteins (SSBs) play essential roles in DNA replication, recombination, and repair in bacteria and eukarya. We report here the identification and characterization of the SSB of an archaeon, Methanococcus jannaschii. The M. jannaschii SSB (mjaSSB) has significant amino acid sequence similarity to the eukaryotic SSB, replication protein A (RPA), and contains four tandem repeats of the core single-stranded DNA (ssDNA) binding domain originally defined by structural studies of RPA. Homologous SSBs are encoded by the genomes of other archaeal species, including Methanobacterium thermoautotrophicum and Archaeoglobus fulgidus. The purified mjaSSB binds to ssDNA with high affinity and selectivity. The apparent association constant for binding to ssDNA is similar to that of RPA under comparable experimental conditions, and the affinity for ssDNA exceeds that for double-stranded DNA by at least two orders of magnitude. The binding site size for mjaSSB is ≈20 nucleotides. Given that RPA is related to mjaSSB at the sequence level and to Escherichia coli SSB at the structural level, we conclude that the SSBs of archaea, eukarya, and bacteria share a common core ssDNA-binding domain. This ssDNA-binding domain was presumably present in the common ancestor to all three major branches of life.
Single-stranded DNA-binding proteins (SSBs) play essential roles in most intracellular transactions involving DNA, including replication, recombination, and repair (1, 2). Members of this class of proteins have been identified in bacterial and eukaryotic cells and also are encoded by mitochondrial and viral genomes. Although there is little detectable amino acid sequence similarity between the bacterial and eukaryotic SSBs, recent evidence suggests these proteins have striking similarities at the structural level and likely are derived from a common ancestor (3–5). No SSB from the third major phylogenetic division, the archaea, has been identified, although recently published sequences of archaeal genomes contain genes whose predicted products have some sequence similarity to replication protein A (RPA), the eukaryotic SSB (6).
RPA originally was purified as a protein essential for SV40 DNA replication in vitro (7–9). Subsequent studies have shown that it is present in cells of all eukaryotic species from yeast to man (1). RPA is a heterotrimer composed of subunits of 70, 32, and 14 kDa. The central 250 amino acids of the large subunit (RPA70) contain two tandem single-stranded DNA-binding domains (3, 10–12). The N terminal region of RPA70 mediates interactions with a variety of cellular proteins, including other replication proteins and certain transcription factors (reviewed in ref. 1). The C terminal region of RPA70 interacts with RPA32 and is required for assembly of the trimeric complex (10–13). The middle subunit of RPA (RPA32) contains an additional single-stranded DNA-binding domain that is likely to be similar in structure to those present in RPA70 (14, 15). It has been suggested on the basis of sequence analysis that the small subunit (RPA14) also may contain a single-stranded DNA-binding domain, but this has not been verified experimentally (14).
A crystal structure at 2.4 Å of the two DNA-binding domains of RPA70 bound to a single-stranded octanucleotide has been reported (3). Each domain is composed of an OB (oligonucleotide/oligosaccharide binding) fold (16) and contains a channel that binds three nucleotides in the DNA chain. The DNA-binding channels of the two domains are oriented so that the single-stranded DNA (ssDNA) extends from one domain to the other in roughly a straight line, with two nucleotides bridging the space between. Both domains interact with the DNA via hydrogen bonds to the bases and phosphate backbone, as well as via stacking interactions between conserved aromatic residues and the bases. Although RPA is the only SSB for which the structure of the protein-DNA complex is known, the structures of Escherichia coli SSB and human mitochondrial SSB in the absence of DNA have significant structural similarity to RPA (3–5, 17).
Here, we report the identification and initial characterization of a SSB from the archaeon Methanococcus jannaschii. We initially identified a putative homologue of RPA70 in the published sequence of the complete M. jannaschii genome (18) and found closely related sequences in the subsequently published genomes of Methanobacterium thermoautotrophicum and Archaeoglobus fulgidus (6, 19). A similar sequence analysis was performed independently by Chedin et al. (20). To determine whether these proteins represent authentic achaeal SSBs, we expressed the putative M. jannaschii SSB in E. coli and purified it to homogeneity. The purified protein possesses single-stranded DNA-binding activity, and its interactions with oligonucleotides are similar in many respects to those of RPA.
MATERIALS AND METHODS
Expression and Purification of M. jannaschii SSB (mjaSSB).
Clone AMJF35, containing ORF MJ1159 of M. jannaschii, was received from the American Type Culture Collection. The MJ1159 ORF was amplified by PCR and was inserted between the BamHI and HindIII sites of the bacterial expression vector pET-28b (Novagen). Transformants of E. coli BL21(DE3) were grown in Luria–Bertani medium and were induced by incubation in the presence of IPTG for 2.5 hr. Bacterial lysates were prepared by sonication in S buffer (50 mM NaH2PO4, pH 8.0/300 mM NaCl/10 mM imidazole) plus 2 μg/ml leupeptin, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml pepstatin, and 2 μg/ml aprotinin. After clarification by centrifugation for 20 min at 13,000 rpm in an Eppendorf centrifuge at 4°C, the lysate from 50–100 ml of culture was mixed with 1–2 ml of Ni-NTA matrix (Qiagen, Chatsworth, CA) for 60 min at 4°C with gentle shaking. The mixture then was poured into a column, was washed with S buffer containing 20 mM imidazole, and was eluted with S buffer containing 250 mM imidazole. The eluate was loaded onto a 1- to 2-ml column of ssDNA agarose (GIBCO/BRL), which was washed with buffer A (25 mM Tris⋅HCl, pH 7.8/1 mM EDTA/10% glycerol/0.01% Tween 20/1 mM DTT/1 mM phenylmethylsulfonyl fluoride) containing 1M NaCl. The protein was eluted from the column with buffer A containing 2 M NaCl/40% ethylene glycol and was dialyzed against buffer A. Protein concentrations were determined by Bradford assay with BSA as standard (21).
Estimation of Native Molecular Weight.
The Stokes radius of mjaSSB was determined by gel filtration of the purified protein at 4°C on a Superdex 200 HR 10/30 FPLC column (Pharmacia) equilibrated with buffer A (without phenylmethylsulfonyl fluoride) containing 0.5 M NaCl. A set of globular protein standards was run separately for reference. The sedimentation coefficient of mjaSSB was determined by density gradient ultracentrifugation of the purified protein mixed with protein standards. The conditions were identical to those used for gel filtration with the exception of the 10–30% glycerol gradient. The native molecular weight of mjaSSB was calculated as described in Siegel and Monty (22).
Gel Mobility Shift Assays.
Oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA) and were end-labeled by using [γ32P]ATP (3,000Ci/mmol) and T4 polynucleotide kinase. Binding reactions were incubated for 45 min at 25°C in HI15 buffer (30 mM Hepes, pH 7.8/1 mM DTT/0.25 mM EDTA/0.5% inositol/15 mM KCl), and the reaction products were resolved by electrophoresis in 1% agarose gels as described (23, 24).
Fluorescence Titrations.
Binding reactions were assembled in 2 ml of FB buffer (30 mM Hepes, pH 7.8/100 mM NaCl/5 mM MgCl2/0.5% inositol/1 mM DTT) and were incubated at either 25 or 50°C. A constant amount of mjaSSB was incubated with varying quantities of poly(dT) (Midland Certified Reagent, Midland, TX). Fluorescence measurements were made on a Jobin-Yvon (Longjumeau, France) Spex FluoroMax-2 spectrofluorometer as described for RPA (23). The excitation wavelength was 295 nm (bandpass 2.5 nm), and the emission wavelength was 348 nm (bandpass 1 nm).
RESULTS
Putative Homologues of RPA in Archaea.
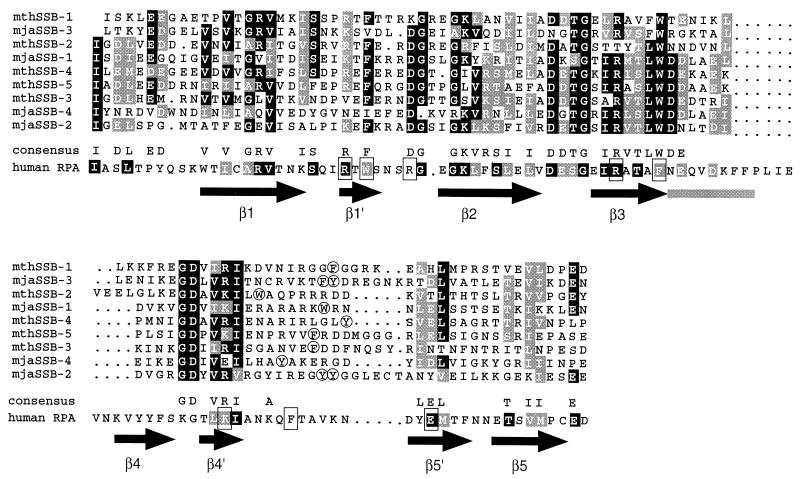
The publication of the first complete sequence of an archaeal genome, M. jannaschii, made it possible to identify candidate DNA replication proteins by sequence similarity searches (18). Although a number of such proteins readily were identified, no homologue of eukaryotic or bacterial SSBs was reported in the initial publication. Given that most M. jannaschii replication proteins appear to be related more closely to their eukaryotic counterparts (see Discussion), we sought to identify putative homologues of human RPA70 (hRPA70). Surveys of the entire M. jannaschii genome with the program fasta (25) revealed an ORF (MJ1159) with low, but perceptible, sequence similarity to hRPA70. More detailed analysis demonstrated that ORF MJ1159 contains four closely related tandem repeats of a sequence encoding ≈100 amino acids. Strikingly, each of the repeats has significant sequence similarity to the first single-stranded DNA-binding domain of hRPA70 (Fig. 1). In many cases, the residues conserved among the four repeats of ORF MJ1159 correspond to those conserved in RPA70 subunits from yeast to man. In particular, most of the amino acids known to contact ssDNA in domain A of human RPA70 (3), including the aromatic residues involved in stacking interactions with the bases, are conserved in the individual repeats of the archaeal protein (Fig. 1). Consistent with these sequence similarities, the predicted secondary structure of the repeats in ORF MJ1159 (26) is compatible with the OB (oligonucleotide/oligosaccharide binding) fold observed in the two tandem DNA-binding domains of RPA70. These findings strongly suggest that the M. jannaschii SSB (mjaSSB) consists of four tandem repeats of a common core ssDNA-binding domain that is structurally similar to that of RPA70. It follows that that the eukaryotic and archaeal SSBs likely evolved from a common ancestor.
Figure 1.
Alignment of the sequences of the ssDNA-binding domains of M. jannaschii and M. thermoautotrophicum SSBs. The four repeats of mjaSSB and the five repeats of mthSSB are numbered sequentially. Identical residues are shaded black, and conserved residues are shaded gray. The sequence of the first ssDNA-binding domain of human RPA70 is shown below the consensus for the archaeal sequences. The shaded residues in the RPA sequence are similar or identical to those of the archaeal consensus. The boxed residues are those that make contact with the ssDNA in the crystal structure of hRPA70 (3). Arrows indicate the positions of the β sheets in the conserved OB (oligonucleotide/oligosaccharide binding) fold of hRPA70, and the gray bar indicates the position of an α helix. Aromatic residues corresponding to the conserved phenylalanine in the in the loop between sheets 4′ and 5′ of RPA70 are circled.
After the analysis described above was completed, the complete sequences of two additional archaeal genomes were published (6, 19). Analysis of these genomes revealed that both contained homologues of mjaSSB. The genome of M. thermoautotrophicum contains an ORF [MT1185/M. thermoautotrophicum SSB (mthSSB)] with five tandem repeats of the core ssDNA-binding domain. Fig. 1 shows the alignment of the nine repeats present in mthSSB and mjaSSB with the sequence of the ssDNA-binding domain of hRPA70. Of interest, the A. fulgidus genome encodes two ORFS (AF0382 and AF0780), each containing two putative ssDNA-binding domains in tandem. This finding may have some evolutionary significance because, as noted above, both the 70-kDa and 32-kDa subunits of hRPA appear to contain core ssDNA-binding domains. Thus, eukaryotes and at least one archaeal species have evolved two separate proteins with presumptive ssDNA-binding activity. It will be of interest to determine whether the two A. fulgidus SSBs exhibit functional differences.
Expression and Purification of the RPA Homologue from M. jannaschii.
To determine whether the ORFs identified by sequence comparison truly represent SSBs, we purified and characterized the candidate SSB from M. jannaschii. The MJ1159 ORF was inserted into the pET-28 bacterial expression vector and was expressed as a fusion protein containing an N-terminal his6 tag. The tagged mjaSSB protein was largely soluble and was purified to near homogeneity by affinity chromatography on Ni-chelate agarose and ssDNA-agarose. Analysis of the purified protein by SDS/PAGE revealed a single major band with a molecular mass of 80 kDa, in agreement with the predicted molecular mass of 77 kDa (data not shown). Analytical gel filtration and sedimentation studies demonstrated that the native mjaSSB has a Stokes radius of 42 Å and a sedimentation coefficient of 4.4 S. The molecular mass of mjaSSB calculated from these values is 76 kDa, indicating that the protein is a monomer in solution.
DNA-Binding Properties of mjaSSB.
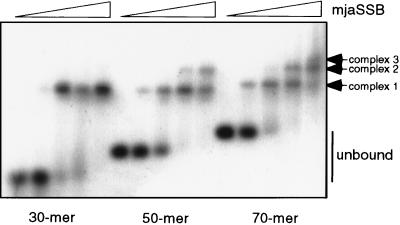
During the purification of the putative mjaSSB, we noted that a high electrolyte concentration in the presence of a chaotropic agent was required to elute the protein from ssDNA agarose. No protein eluted from the column at 1M NaCl, but quantitative recovery was obtained by elution with 2 M NaCl/40% ethylene glycol. To confirm the ability of mjaSSB to bind to ssDNA, we carried out agarose gel mobility shift assays with radioactively labeled single-stranded dT-oligonucleotides 30, 50, and 70 nucleotides in length (Fig. 2). When (dT)30 was incubated with increasing concentrations of mjaSSB, a single band of reduced mobility was observed. Most of the (dT)30 was shifted after addition of one molar equivalent of mjaSSB, and the mobility of the shifted band remained constant at higher protein concentrations. A band of identical mobility was observed for (dT)50 at low oligonucleotide concentrations, but a second shifted band with a lower mobility was observed at high protein concentrations. In the case of (dT)70, a third shifted band appeared at the highest protein concentrations. Assuming that mjaSSB is a monomer, the simplest interpretation of these data is that a single protein molecule binds to (dT)30 whereas two and three protein molecules, respectively, bind to (dT)50 and (dT)70. Under similar experimental conditions, a single molecule of hRPA binds to (dT)30 and (dT)50 whereas two molecules of hRPA bind to (dT)70 (23, 24). Thus, the binding site size for mjaSSB may be somewhat smaller than that of hRPA.
Figure 2.
Binding of mjaSSB to single-stranded oligonucleotides. A fixed quantity of [5′-32P]oligonucleotide (38 fmol of dT30, 32 fmol of dT50, and 32 fmol of dT70) was incubated with 0, 5, 20, 50, and 200 fmol of mjaSSB in 20-μl reaction mixtures at 25°C. Protein–DNA complexes were separated from free DNA by agarose gel electrophoresis.
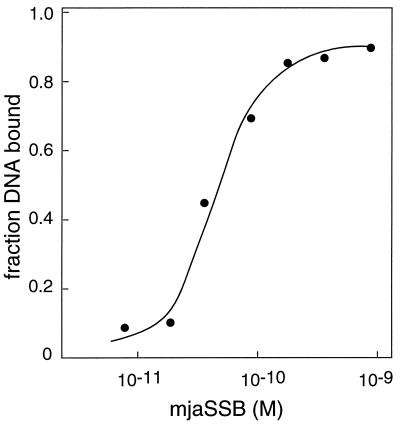
To determine the apparent affinity of mjaSSB for ssDNA, radioactively labeled (dT)30 was incubated with increasing quantities of protein, and the fraction of DNA bound was measured by agarose gel mobility shift assay as before (Fig. 3). The apparent binding constant of mjaSSB for (dT)30 was on the order of 2 × 1010 M−1. The apparent affinity of hRPA for ssDNA measured by the same experimental method is ≈1 × 1010 M−1 (23, 24, 27).
Figure 3.
Binding isotherm for mjaSSB with (dT)30. Binding reaction mixtures (20 μl) contained 0.03 fmol of [5′-32P]dT30 and the indicated concentration of mjaSSB. Protein–DNA complexes were separated from free DNA by agarose gel electrophoresis, and the radioactivity in each species was determined by using a phosphorimager. The apparent Ka is 2 × 1010 M−1.
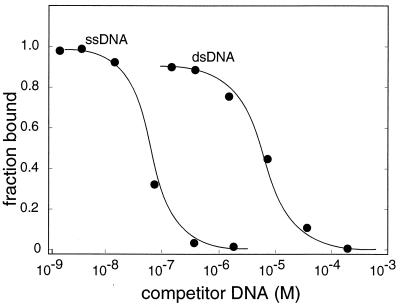
To compare the affinity of mjaSSB for single- and double-stranded DNA, we performed binding competition studies. Reaction mixtures containing fixed amounts of mjaSSB and radioactive (dT)30 were incubated with increasing concentrations of unlabeled single- or double-stranded M13 DNA (Fig. 4). In the absence of competitor, the concentration of mjaSSB was sufficient to drive most of the (dT)30 into protein–DNA complexes. Approximately 100× as much double-stranded competitor DNA as single-stranded competitor DNA was required to reduce the fraction of (dT)30 in protein–DNA complexes to 50%. These data indicate that the apparent affinity of mjaSSB for ssDNA is at least 100× that for double-stranded DNA. This estimate probably represents a lower limit because preparations of double-stranded M13 DNA likely contain low levels of contamination with ssDNA.
Figure 4.
Relative affinity of mjaSSB for single- and double-stranded DNA. Reaction mixtures (20 μl) contained a fixed quantity of mjaSSB (1.75nM) and [32P]dT30 (0.05nM). Before incubation, the [32P]dT30 was mixed with single- or double-stranded M13 DNA to the indicated final concentration. Protein–DNA complexes were separated from free DNA by agarose gel electrophoresis, and the fraction of DNA bound to protein was estimated by quantification of digitized images of scanned autoradiograms.
Binding Site Size of mjaSSB.
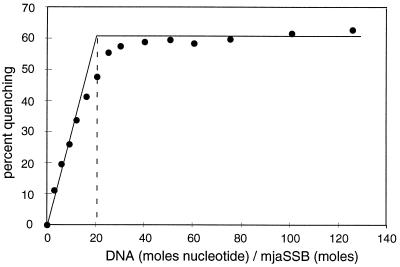
To further explore the binding properties of mjaSSB, we made use of fluorescence spectroscopy. The mjaSSB protein contains six tryptophan residues, and, based on the alignment of the mjaSSB repeats with the sequence of RPA, five of the six residues are likely to be located in DNA binding sites (see Fig. 1). We observed that the intrinsic fluorescence of mjaSSB, like that of RPA, is quenched on DNA binding (23). With an excitation wavelength of 295 nm, the emission spectrum of mjaSSB at 25°C had a maximum at 348 nm, consistent with tryptophan fluorescence. On addition of a saturating quantity of ssDNA, the intrinsic fluorescence at 348 nm was reduced (quenched) by ≈60% (Fig. 5). DNA-dependent quenching also was observed at 50°C (data not shown), indicating that mjaSSB is active in DNA binding at high temperature.
Figure 5.
Binding site size of mjaSSB. Reaction mixtures (2 ml) contained a fixed amount of mjaSSB (0.4 nmol). The indicated amounts of poly(dT) were added, and the fluorescence, at 348 nm was measured at 25°C with an excitation wavelength of 295 nm. The percent quenching is the change in mjaSSB fluorescence normalized to the initial fluorescence in the absence of poly(dT).
The occluded binding site size of a SSB is the length of DNA rendered inaccessible when the protein is bound. To estimate this binding parameter for mjaSSB, we used fluorescence measurements to follow binding of the protein to DNA under stoichiometric binding conditions (Fig. 5). When a fixed quantity of mjaSSB was titrated with poly(dT), the quenching of mjaSSB fluorescence increased linearly with DNA concentration until saturation was reached (Fig. 5). At saturation, the molar ratio of nucleotide to protein was 21. This binding site size is somewhat smaller than that of RPA, which was estimated to be 30 nucleotides by similar methodology (23).
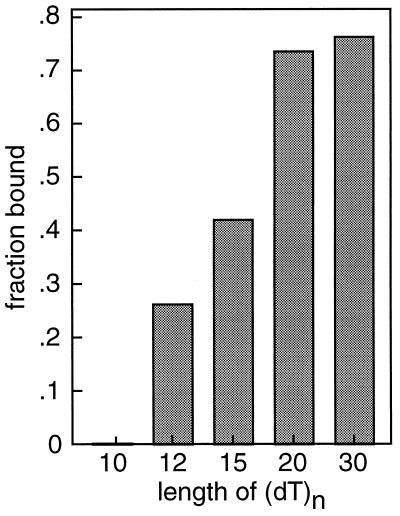
We also studied the interaction of mjaSSB with oligonucleotides of different lengths by using gel mobility shift assays. Fig. 6 shows the results of an experiment in which a fixed quantity of mjaSSB was incubated with radioactively labeled (dT)n oligonucleotides ranging in length from 10 to 30 nucleotides. The molar concentration of oligonucleotide added to each reaction mixture was the same, so that the relative amount of protein–oligonucleotide complex formed in each case represented a measure (albeit qualitative) of the relative affinity of binding to mjaSSB. Under the conditions of this experiment, about three-fourths of the (dT)30 was shifted to the position of protein–DNA complexes. A slightly smaller amount of protein–DNA complexes was observed for (dT)20. For oligonucleotides with lengths <20, the binding of mjaSSB showed a strong dependence on length, and complexes between (dT)10 and mjaSSB were not detectable at all (although such complexes could be observed at higher protein concentrations). These data are consistent with a binding site size of 15–20 nucleotides for mjaSSB.
Figure 6.
Interaction of mjaSSB with oligonucleotides of different lengths. Reaction mixtures (20 μl) contained 1 fmol of oligonucleotide of the indicated length and 5 fmol of mjaSSB. After incubation at 25°C for 30 min, protein–DNA complexes were separated from free DNA by agarose gel electrophoresis. The fraction of DNA bound to protein was estimated by quantitation of digitized images of scanned autoradiograms.
DISCUSSION
When the first complete sequence of an archaeal genome was published, it became apparent that the predicted amino acid sequences of archaeal DNA replication proteins more closely resemble those of eukaryotes than bacteria (Table 1) (18, 28). This is consistent with a large body of evidence indicating that the divergence of archaea and eukaryotes is more recent than the divergence of bacteria and eukaryotes/archaea (29). Archaea and eukaryotes contain some replication proteins for which no bacterial homologue is yet evident. This is especially striking for proteins thought to be involved in initiation of DNA replication in eukaryotic cells, such as members of the Orc1/Cdc18/Cdc6 and MCM families of initiation proteins. In other cases, it is possible to identify a likely bacterial homologue whose amino acid sequence is distantly related to the corresponding eukaryotic/archaeal replication protein. For example, there is weak, but significant, primary sequence similarity between the proteins of bacteria and eukarya/archaea that are responsible for loading processivity factors at the replication fork (“clamp-loading complexes”). The similarity of both sequence and function suggests that these proteins evolved from a common ancestor (28). Even in cases in which no amino acid sequence similarity can be detected between a eukaryotic/archaeal replication protein and its bacterial counterpart, a likely evolutionary relationship can be revealed by structural studies. Thus, the processivity factor of eukaryotes, PCNA, shows no sequence similarity to the β clamp of E. coli, but the three dimensional structures of the two proteins are superimposible (30).
Table 1.
Homologues of eukaryotic DNA replication proteins
| Eukaryotic protein | Functional role | Archaeal homologue | Bacterial homologue |
|---|---|---|---|
| Orc1/Cdc18/Cdc6 | Initiation | Yes | ? |
| MCM complex | Initiation | Yes | ? |
| RPA (ssDNA binding) | Initiation/elongation | Yes | Yes |
| Primase | Initiation/elongation | Yes | ? |
| DNA polymerase (B family) | Elongation | Yes | Yes |
| RFC (clamp loader) | Elongation | Yes | Yes |
| PCNA | Elongation | Yes | Yes |
| DNA ligase | Elongation | Yes | Yes |
| FENI | Elongation | Yes | ? |
| Ribonuclease H | Elongation | Yes | Yes |
| Dna2 helicase | Elongation | Yes | ? |
| Topoisomerase I | Elongation | Yes | Yes |
Entries in the table are derived in part from ref. 28 and in part from blast searches with the archaeal and/or eukaryotic replication protein as query. See also ref. 6, 18, and 19 and the web sites http://www.tigr.org/tdb/mdb/mjdb/mjdb.html, http://www.tigr.org/tdb/mdb/afdb/afdb.html, and http://www.biosci.ohio-state.edu/∼genomes/mthermo/.
Our data indicate that the archaeal SSB has detectable primary sequence similarity to eukaryotic RPA but not to E. coli SSB. Although it has been suggested that that E. coli SSB and RPA share amino acid sequence similarity (14), the proposed sequence alignment is very weak and is inconsistent with the available structural data (see below). Thus, on the basis of sequence data alone, one cannot conclude that the archaeal/eukaryotic SSBs and the bacterial SSBs have a common origin. However, comparison of the three-dimensional structures of the ssDNA-binding domains of RPA and E. coli SSB makes a compelling case for an evolutionary relationship between the two proteins. The alignment of the first ssDNA-binding domain of RPA with E. coli SSB reveals a rms deviation of 2.2 Å for the equivalent C-α atoms over a length of 72 amino acid residues (Biomolecule 3D Structure data base, National Center for Biotechnology Information; see ref. 31). The probability of this structural similarity arising by chance is 10−5. Thus, it is likely that the SSBs of modern organisms share a single core ssDNA-binding domain that was already present in the common ancestor to bacteria, archaea, and eukarya. We suggest that most, if not all, of the protein functional units at the replication fork are of similarly ancient origin. With respect to the proteins specifically involved in initiation, it is not yet clear whether novel functional units evolved in the lineage that gave rise to the archaea and eukarya or whether there is simply insufficient data to allow recognition of possible bacterial homologues.
The purified mjaSSB has high affinity and selectivity for ssDNA. The apparent association constant is similar to that of RPA under comparable experimental conditions. Although we have not studied the temperature dependence of DNA binding by mjaSSB in detail, our fluorescence quenching data indicate that the protein binds ssDNA at high temperatures. M. jannaschii grows at an optimum temperature of 85°C at pressures up to 200 atm (18). Thus, it will be of considerable interest to explore further the interaction of mjaSSB with oligonucleotides under more extreme conditions.
The occluded binding site size of mjaSSB is in the neighborhood of 20 nucleotides. This value is consistent with expectations based on analysis of the structure of human RPA. The ssDNA-binding channel of each domain of hRPA70 accommodates three nucleotides, and the space between the two domains is occupied by two nucleotides (3). Because mjaSSB has four tandem ssDNA-binding domains, an occluded binding site size of at least 12–20 nucleotides is predicted. In addition to the four DNA-binding domains, mjaSSB, like other archaeal SSBs, possesses a putative zinc finger motif near the C terminus (20). RPA70 contains a similar motif, but genetic studies indicate that it is not required for ssDNA binding (1).
It is interesting that mjaSSB contains four tandem repeats of the conserved core ssDNA-binding domain. The homologues of mjaSSB in other archaeal species also are organized as tandem repeats. The SSB of M. thermoautotrophicum contains five repeats of the core ssDNA-binding domain. The A. fulgidus genome encodes two different SSBs, each of which contains two such repeats. As noted above, it has been suggested on the basis of sequence comparisons that the RPA may contain as many as four core ssDNA-binding domains distributed over the three subunits (14). Three of these have been verified by biochemical studies (1, 15). The E. coli SSB contains only a single core ssDNA-binding domain, but the protein functions as a homotetramer (2). Thus, the functional form of all SSBs appears to consist of several core ssDNA-binding domains, but these domains can be associated with one or more distinct polypeptide chains. This organization may impart considerable flexibility to the interactions of SSBs with ssDNA. Indeed, multiple modes of protein–DNA interaction have been reported for both RPA and E. coli SSB (1, 2). The modular structure of SSBs also may function to promote very tight binding to ssDNA while at the same time allowing the rapid stepwise removal of the proteins from the DNA as must occur during DNA chain elongation on a ssDNA template.
The mjaSSB protein appears to be considerably less complex than RPA70. In particular, it lacks the N- and C-terminal regions through which RPA70 interacts with a variety of other cellular proteins. The latter domains of RPA70 likely evolved after the divergence of archaea and eukarya as a response to the more complex regulatory requirements of replicating very large genomes with multiple origins of replication. In addition, the published genomes of archaeal species do not reveal sequences with similarity to RPA32 and RPA14 subunits of RPA, so it is possible that mjaSSB functions as a single polypeptide chain. Thus, mjaSSB may represent a simple model for studying the structure, function, and evolution of the conserved core ssDNA-binding domain.
Acknowledgments
We thank members of the Kelly lab for thoughtful discussions. This work was supported by research grants from the National Institutes of Health.
ABBREVIATIONS
- SSB
single-stranded DNA-binding protein
- RPA
replication protein A
- ssDNA
single-stranded DNA
- mjaSSB
Methanococcus jannaschii SSB
- mthSSB
Methanobacterium thermoautotrophicum SSB
References
- 1.Wold M S. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 2.Lohman T M, Ferrari M E. Annu Rev Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 3.Bochkarev A, Pfuetzner R A, Edwards A M, Frappier L. Nature (London) 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 4.Raghunathan S, Ricard C S, Lohman T M, Waksman G. Proc Natl Acad Sci USA. 1997;94:6652–6657. doi: 10.1073/pnas.94.13.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webster G, Genschel J, Curth U, Urbanke C, Kang C, Hilgenfeld R. FEBS Lett. 1997;411:313–316. doi: 10.1016/s0014-5793(97)00747-3. [DOI] [PubMed] [Google Scholar]
- 6.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, et al. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wobbe C R, Weissbach L, Borowiec J A, Dean F B, Murakami Y, Bullock P, Hurwitz J. Proc Natl Acad Sci USA. 1987;84:1834–1838. doi: 10.1073/pnas.84.7.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wold M S, Kelly T J. Proc Natl Acad Sci USA. 1988;85:2523–2527. doi: 10.1073/pnas.85.8.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fairman M P, Stillman B. EMBO J. 1988;7:1211–1218. doi: 10.1002/j.1460-2075.1988.tb02933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomes X V, Wold M S. J Biol Chem. 1995;270:4534–4543. doi: 10.1074/jbc.270.9.4534. [DOI] [PubMed] [Google Scholar]
- 11.Gomes X V, Wold M S. Biochemistry. 1996;35:10558–10568. doi: 10.1021/bi9607517. [DOI] [PubMed] [Google Scholar]
- 12.Lin Y L, Chen C, Keshav K F, Winchester E, Dutta A. J Biol Chem. 1996;271:17190–17198. doi: 10.1074/jbc.271.29.17190. [DOI] [PubMed] [Google Scholar]
- 13.Kim D K, Stigger E, Lee S H. J Biol Chem. 1996;271:15124–15129. doi: 10.1074/jbc.271.25.15124. [DOI] [PubMed] [Google Scholar]
- 14.Philipova D, Mullen J R, Maniar H S, Lu J, Gu C, Brill S J. Genes Dev. 1996;10:2222–2233. doi: 10.1101/gad.10.17.2222. [DOI] [PubMed] [Google Scholar]
- 15.Bochkareva E, Frappier L, Edwards A M, Bochkarev A. J Biol Chem. 1998;273:3932–3936. doi: 10.1074/jbc.273.7.3932. [DOI] [PubMed] [Google Scholar]
- 16.Murzin A G. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang C, Curth U, Urbanke C, Kang C. Nat Struct Biol. 1997;4:153–157. doi: 10.1038/nsb0297-153. [DOI] [PubMed] [Google Scholar]
- 18.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 19.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, et al. Nature (London) 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 20.Chedin F, Seitz E M, Kowalczykowski S C. Trends Biochem Sci. 1998;23:273–277. doi: 10.1016/s0968-0004(98)01243-2. [DOI] [PubMed] [Google Scholar]
- 21.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 22.Siegel L M, Monty K J. Biochim Biophys Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- 23.Kim C, Paulus B F, Wold M S. Biochemistry. 1994;33:14197–206. doi: 10.1021/bi00251a031. [DOI] [PubMed] [Google Scholar]
- 24.Kim C, Snyder R O, Wold M S. Mol Cell Biol. 1992;12:3050–3059. doi: 10.1128/mcb.12.7.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pearson W R, Lipman D J. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chou P Y, Fasman G D. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 27.Kim C, Wold M S. Biochemistry. 1995;34:2058–2064. doi: 10.1021/bi00006a028. [DOI] [PubMed] [Google Scholar]
- 28.Edgell D R, Doolittle F W. Cell. 1997;89:995–998. doi: 10.1016/s0092-8674(00)80285-8. [DOI] [PubMed] [Google Scholar]
- 29.Olsen G J, Woese C R. Cell. 1997;89:991–994. doi: 10.1016/s0092-8674(00)80284-6. [DOI] [PubMed] [Google Scholar]
- 30.Kelman Z, O’Donnell M. Nucleic Acids Res. 1995;23:3613–3620. doi: 10.1093/nar/23.18.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gibrat J-F, Madej T, Bryant S H. Curr Opin Struct Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]