Abstract
Assembly of several inner membrane proteins—leader peptidase (Lep), a Lep derivative (Lep-inv) that inserts with an inverted topology compared with the wild-type protein, the phage M13 procoat protein, and a procoat derivative (H1-procoat) with the hydrophobic core of the signal peptide replaced by a stretch from the first transmembrane segment in Lep—has been studied in vitro and in Escherichia coli strains that are conditional for the expression of either the 54 homologue (Ffh) or 4.5S RNA, which are the two components of the E. coli signal recognition particle (SRP), or SecE, an essential core component of the E. coli preprotein translocase. Membrane insertion has also been tested in a SecB null strain. Lep, Lep-inv, and H1-procoat require SRP for correct assembly into the inner membrane; in contrast, we find that wild-type procoat does not. Lep and, surprisingly, Lep-inv and H1-procoat fail to insert properly when SecE is depleted, whereas insertion of wild-type procoat is unaffected under these conditions. None of the proteins depend on SecB for assembly. These observations indicate that inner membrane proteins can assemble either by a mechanism in which SRP delivers the protein at the preprotein translocase or by what appears to be a direct integration into the lipid bilayer. The observed change in assembly mechanism when the hydrophobicity of the procoat signal peptide is increased demonstrates that the assembly of an inner membrane protein can be rerouted between different pathways.
Keywords: preprotein translocase/protein targeting
What are the mechanisms that ensure efficient targeting and assembly of integral membrane proteins in various organisms and organelles? In eukaryotic cells, the biosynthesis of almost all endoplasmic reticulum (ER) membrane proteins seems to proceed by the same mechanism (1–3). The process starts with the signal recognition particle (SRP) binding to the N-terminal signal or signal-anchor sequence when the nascent chain has reached a critical length of approximately 60 amino acids (i.e., when the signal sequence is exposed just outside the ribosome). Further translation is inhibited until the SRP contacts its receptor at the ER membrane and dissociates from the nascent chain. The ribosome then makes a tight seal with the translocon (Sec61p complex), translation is resumed, and the nascent chain inserts cotranslationally into the aqueous translocation channel. Hydrophilic polypeptide chains are translocated across the ER membrane through the translocon, whereas hydrophobic transmembrane segments get trapped in the translocon and subsequently move out laterally into the lipid bilayer (4).
Homologues of the eukaryotic SRP, its receptor, and most of the translocon subunits are present in Escherichia coli (1, 5). Recently, evidence has emerged that the E. coli SRP-targeting pathway may be involved in the assembly of at least a subset of inner membrane proteins (6–12). However, the role of the E. coli Sec machinery (which includes the SecB chaperone; the preprotein translocase, including the integral membrane subunits SecY, SecG, and SecE; and the peripheral subunit SecA) in the assembly of inner membrane proteins is not clear. It has been proposed that inner membrane proteins, except for those containing long periplasmic loops, may assemble in a Sec-independent fashion (13). It should be noted, however, that this conclusion was based on negative results from either conditional secA and secY strains that were primarily selected for secretion defects rather than inner membrane protein assembly defects or by sodium azide treatment that only partially inhibits the ATPase activity of the preprotein translocase subunit SecA (14, 15).
To further delineate the role of the SRP-targeting pathway and the preprotein translocase in the assembly of inner membrane proteins in E. coli, we have studied four model inner membrane proteins with different membrane topologies and different modes of membrane assembly both in vitro and in vivo in various depletion backgrounds. Leader peptidase (Lep) has two transmembrane segments and a large C-terminal periplasmic domain (Fig. 1). Translocation of the C-terminal domain depends on SecA and SecY (19), but translocation of the N-terminal tail does not (20). Lep-inv, the second model protein, is a Lep derivative with an inverted membrane topology where the short P1 loop is exposed to the periplasm; translocation of the P1 loop appears to be independent of SecA and SecY (21). Both Lep and Lep-inv require the E. coli SRP for efficient targeting to the inner membrane (7). The phage M13 procoat protein has a cleavable N-terminal signal peptide, a short periplasmic loop, and a short cytoplasmic C-terminal tail. It does not appear to require SecA or SecY for assembly (19, 22, 23). Its dependence on the E. coli SRP has not previously been tested. Finally, we have constructed a modified procoat (H1-procoat), where the core of the signal peptide has been replaced by a more hydrophobic stretch from the first transmembrane segment of Lep.
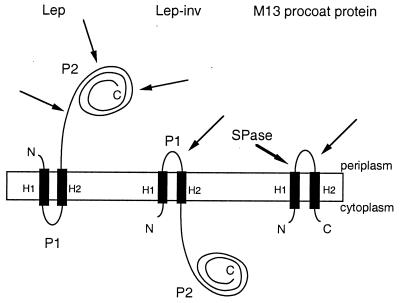
Figure 1.
Orientation of Lep, Lep-inv, and M13 procoat protein in the cytoplasmic membrane. The Lep-inv mutant was derived from Lep by adding 3 lysine codons between codons 4 and 5, inserting 10 codons encoding the sequence Gly-Gln-Ser-Leu-Asn-Ala-Pro-Thr-Ser-Gly between codons 22 and 24, deleting residues 30–52, and changing Lys56 to Asn and Glu61 to Val (16, 17). In spheroplasts, proteinase K degrades the periplasmic P2 domain of Lep and the periplasmic P1 loop of Lep-inv (thin arrows). For Lep-inv, this treatment gives rise to a protease-resistant H2-P2 fragment that can be immunoprecipitated with a Lep antiserum, whereas no immunoprecipitable material remains when the P2 domain in Lep has been digested. M13 procoat protein is processed by signal peptidase I (SPase; thick arrow) and can be immunoprecipitated with an antiserum raised against the periplasmic domain of the mature coat protein. In spheroplasts, proteinase K degrades the periplasmic domain of M13 coat protein and thus abolishes the immunoreactivity of the protein (18).
Membrane assembly of Lep, Lep-inv, procoat, and H1-procoat have been studied in Escherichia coli strains that are conditional for the expression of either 54 homologue (Ffh) or 4.5S RNA, the two components of the E. coli SRP, or SecE, an essential component of the E. coli preprotein translocase (1, 24, 25). In addition, membrane assembly of the model proteins has been studied in a SecB null mutant strain. SecB is a chaperone that is involved in posttranslational targeting (24, 26), and it has been suggested that proteins that use the SRP pathway do not use SecB and vice versa (10).
MATERIALS AND METHODS
Strains, Plasmids, and Growth Conditions.
The 4.5S RNA conditional strain FF283 was cultured as described (27). To deplete cells of 4.5S RNA, cells were grown to midlogarithmic phase in the absence of isopropyl β-d-thiogalactopyranoside (IPTG). The Ffh conditional strain WAM121 (7) was cultured in LB medium supplemented with 0.4% glucose and 0.1% l-arabinose. Overnight cultures were washed once with LB and back-diluted 1:20. To deplete cells for Ffh, cells were grown to midlogarithmic phase in the absence of l-arabinose. Thirty minutes before labeling the WAM121 cells were transferred to M9 minimal medium. SRP depletion was checked by monitoring the accumulation of pre-β-lactamase during a short pulse labeling with [35S]methionine. The SecB null mutant strain MM152 (28) was cultured in Mops medium (29). The SecE-depletion strain CM124 (30) was cultured in M9 minimal medium supplemented with 0.4% glucose and 0.2% l-arabinose. Overnight cultures were washed once with M9 medium and back-diluted 1:20. To deplete cells for SecE, cells were grown to midlogarithmic phase in the absence of l-arabinose. Depletion of SecE was checked by monitoring the accumulation of pro-outer membrane protein A (OmpA) during a short pulse labeling with [35S]methionine. Where appropriate, ampicillin (final concentration, 100 μg/ml) and kanamycin (final concentration, 50 μg/ml) were added to the medium.
Lep, Lep-inv, M13 procoat, and H1-procoat (a procoat derivative in which the underlined residues in the wild-type signal sequence MKKSLVLKASVAVATLVPMLSFA have been replaced by a more hydrophobic stretch from the H1 transmembrane segment in Lep; MKKSLVLFALILVIPMLSFA) were expressed by l-arabinose induction from the pING1 vector (31) in the strains FF283 and MM152 and by IPTG induction from the pDHB5700 vector (unpublished results, a gift from J. Beckwith, Harvard Medical School, Boston) in strain CM124. In strain WAM121, proteins were expressed from the pJF119HE vector by IPTG induction (32).
Assay for Membrane Targeting.
For all experiments, cells were grown to midlogarithmic phase. Expression of Lep, Lep-inv, M13 procoat protein, and H1-procoat protein was induced for 5 min with either IPTG (final concentration, 1 mM) or l-arabinose (final concentration, 0.2%). Cells were labeled with [35S]methionine (150 μCi/ml; 1 Ci = 37 GBq) for 15 sec, and then nonradioactive methionine was added (final concentration, 500 μg/ml). FF283 and WAM121 cells expressing M13 procoat protein and H1-procoat protein were directly precipitated with trichloroacetic acid (final concentration, 10%), resuspended in 10 mM Tris/2% SDS, and processed as described below. Otherwise, cells were converted to spheroplasts. For spheroplasting, cells were collected at 14,000 rpm for 2 min in a microcentrifuge, resuspended in ice-cold buffer [40% (wt/vol) sucrose/33 mM Tris⋅HCl, pH 8.0), and incubated with lysozyme (final concentration, 5 μg/ml) and 1 mM EDTA for 15 min on ice. Aliquots of the spheroplast suspension were incubated on ice for 1 h either in the presence or absence of proteinase K (final concentration, 0.3 mg/ml). Subsequently, phenylmethylsulfonyl fluoride was added to the spheroplast suspensions (final concentration, 0.33 mg/ml). After addition of phenylmethylsulfonyl fluoride, samples were precipitated with trichloroacetic acid (final concentration, 10%), resuspended in 10 mM Tris/2%SDS, immunoprecipitated with antiserum to Lep, M13 coat protein, OmpA [a periplasmic control (7)], AraB/bandX [a cytoplasmic control (7), results not shown], and β-lactamase (a control to monitor SRP depletion), washed, and analyzed by gel electrophoresis. M13 procoat protein and H1-procoat protein were analyzed by Tricine SDS/PAGE (33), and all other proteins were analyzed by standard SDS/PAGE (34). Gels were scanned and quantitated in a Fuji BAS1000 phosphoimager.
In Vitro Transcription, Translation, Targeting, Cross-Linking, and Floatation Gradient Analysis.
In vitro transcription to produce truncated mRNAs that carry a C-terminal methionine tag sequence was done with the pC4Meth-vector as described by Valent et al. (11). Translation, targeting, cross-linking, and floatation gradient analysis were essentially performed as described by Valent et al. (12). The M13 procoat nascent chains that were used in the in vitro studies contained the 73 amino acids of the full-length M13 procoat and the H1-procoat nascent chains that were used in the in vitro studies contained the 71 amino acids of the full-length H1-procoat protein.
Hydrophobicity Calculations.
The mean hydrophobicity of the (Ala, Leu)-based PhoA signal sequences from Doud et al. (35), procoat, and H1-procoat was calculated by using the toppred program (36) and the GES hydrophobicity scale (37). The full and core window lengths were set to 11 and 9 residues, respectively. Procoat has a lysine residue in the h-region of the signal sequence (K8) that reduces the calculated mean hydrophobicity of the h-region (residues 4–14) to a very low value (〈H〉 = 1.4); however, if the positively charged moiety on the lysine side chain is allowed to “snorkel” (38) along the h-region, the aliphatic part of the side chain will in fact contribute to the overall hydrophobicity. For this reason, the calculation was repeated with K8 changed to either Ala or Leu, and the respective mean hydrophobicities for the h-region were found to be 〈H〉 = 2.0 and 2.1.
RESULTS
M13 Procoat Protein Does Not Depend on the E. coli SRP for Efficient Insertion into the Inner Membrane.
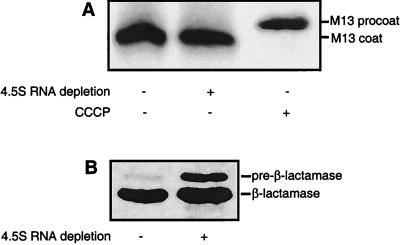
We have previously shown, by using a pulse–chase/protease accessibility protocol, that both Lep and Lep-inv require the E. coli SRP for correct assembly into the inner membrane (7). It has been proposed, however, that not all inner membrane proteins depend on the SRP-targeting pathway for assembly (9). We decided to test the SRP dependency of the M13 procoat protein, which is thought to insert spontaneously into the inner membrane (22, 39, 40). As seen in Fig. 2, the assembly of procoat (as indicated by the removal of the signal peptide) was unaffected upon depletion of the 4.5S RNA. Similarly, no effect on membrane insertion was seen in the Ffh depletion strain WAM121 (data not shown). Thus, in contrast to both Lep and Lep-inv, procoat appears not to be targeted by the SRP pathway. As reported (18), dissipation of the membrane potential by addition of the protonophore carbonyl cyanide m-chlorophenylhydrazone efficiently inhibited membrane insertion, leading to accumulation of the pro-form of the protein.
Figure 2.
SRP is not required for efficient membrane assembly of procoat. (A) Proper membrane assembly is shown by the rapid processing of the procoat signal peptide in strain FF283 not depleted (lane 1) or depleted (lane 2) for the 4.5S RNA. In lane 3, carbonyl cyanide m-chlorophenylhydrazone was added to inhibit insertion (and subsequent processing) of procoat (18) in FF283 cells not depleted for the 4.5S RNA. (B) Depletion of the 4.5S RNA in strain FF283 was checked by monitoring the accumulation of pre-β-lactamase (27). Lanes: 1, strain FF283 not depleted for the 4.5S RNA; 2, strain FF283 depleted for the 4.5S RNA.
Lep and Lep-inv Depend on a Functional Preprotein Translocase for Efficient Insertion into the Inner Membrane, Whereas M13 Procoat Does Not.
In eukaryotic cells, the SRP-targeting pathway delivers membrane proteins at the translocon (Sec61p complex). The E. coli SRP and preprotein translocase and eukaryotic SRP and translocon components are strongly related (1, 5). The core of the E. coli preprotein translocase consists of SecE and SecY, which are homologous to the eukaryotic translocon core subunits Sec61γ and Sec61α, respectively (1), and SecG and SecA, which have no known eukaryotic ER translocon homologues (1). In E. coli, the depletion of SecE results in a sharp decrease in the levels of all components of the preprotein translocase but SecA (25), and it has been shown that SecY is degraded by the FtsH protease in the absence of SecE (41). SecE depletion is the most effective way to compromise the E. coli preprotein translocase in vivo found so far, and precursor forms of exported proteins such as DegP, whose secretion is not noticeably affected in secA and secY conditional strains, can be readily observed in the SecE-depletion strain CM124 (30).
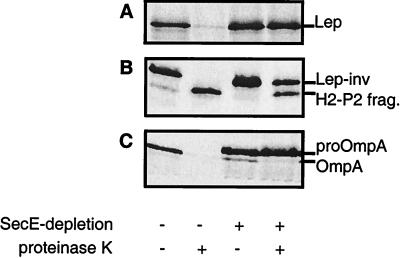
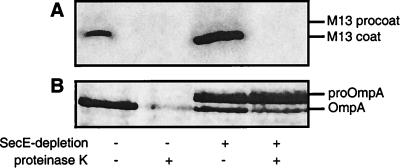
Although previous studies have failed to demonstrate a dependence on SecA and SecY for the assembly of Lep-inv and procoat (19, 21), the availability of the SecE-depletion strain CM124 prompted us to reinvestigate this issue. Upon depletion of SecE, the assembly of the SecA- and SecY-dependent Lep protein was, as expected, strongly affected (Fig. 3A). Unexpectedly, however, the assembly of Lep-inv was also strongly affected under these conditions (Fig. 3B). In contrast to the assembly of Lep and Lep-inv, the assembly of the SRP-independent procoat protein was not detectably affected by SecE depletion (Fig. 4).
Figure 3.
Lep and Lep-inv depend on a functional preprotein translocase for efficient insertion into the cytoplasmic membrane. Lep (A) and Lep-inv (B) were expressed in strain CM124 not depleted (lanes 1 and 2) and depleted (lanes 3 and 4) for SecE. Cells were pulse-labeled for 15 sec and subsequently processed. (B) The H2-P2 product is a protease-resistant fragment resulting from cleavage in the P1 loop (Fig. 1). OmpA secretion/processing was monitored in parallel to check spheroplasting and SecE depletion (C).
Figure 4.
M13 procoat protein does not depend on a functional preprotein translocase for efficient insertion into the cytoplasmic membrane. (A) Procoat was expressed in strain CM124 not depleted (lanes 1 and 2) and depleted (lanes 3 and 4) for SecE. Cells were pulse-labeled for 15 sec and subsequently processed. (B) OmpA secretion/processing was monitored in parallel to check spheroplasting and SecE depletion.
M13 Procoat Associates with Inner Membranes Only When Released from the Ribosome and Does Not Interact with the SRP and Preprotein Translocase.
In vitro floatation assay studies have shown that ribosome-bound Lep nascent chains associate with inner membranes, and cross-linking studies have further shown that these Lep nascent chains interact with the SRP in the cytosol and are transferred to the SecA and SecY components of the preprotein translocase in the membrane after the release of the SRP from the nascent chains by the SRP receptor FtsY (12). The in vivo results reported above are thus in full agreement with the in vitro data.
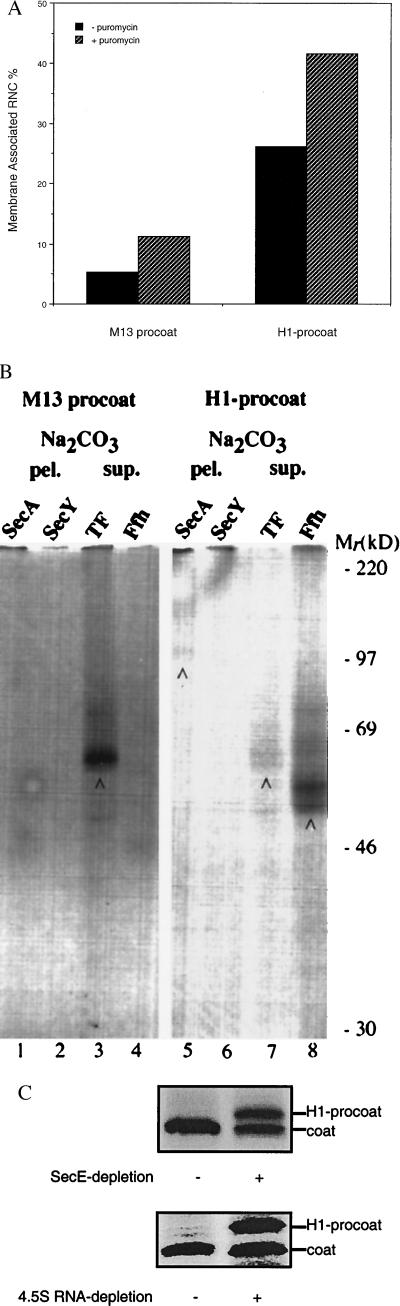
Procoat targeting and assembly have not been studied previously by using an in vitro cross-linking approach. Therefore, ribosome-nascent chain complexes were prepared by translating mRNAs encoding the full-length M13 procoat, in an E. coli cell-free extract (11); these ribosome-bound nascent chains are of sufficient length (77 amino acids) to expose the signal sequence to the outside of the ribosome, assuming that 30–40 amino acids are sequestered in the ribosome. Purified ribosome–nascent chain complexes were incubated with inner membrane vesicles and subjected to floatation gradient analysis (12) to study the interaction of ribosome-bound procoat nascent chains with the inner membrane. As shown in Fig. A, only background levels of membrane-associated (floated) ribosome–nascent chain complexes were observed. When the nascent chains were released from the ribosomes with puromycin before the addition of membranes, a weak but reproducible association of nascent chains with inner membranes could be detected.
To probe the molecular environment of ribosome-bound procoat nascent chains, a cross-linking approach was used (12). Nascent polypeptides were incubated with inner membrane vesicles to allow targeting and subsequently treated with the bifunctional cross-linking reagent disuccinimidyl suberate. Disuccinimidyl suberate is a membrane-permeable analogue of BS3, a cross-linking reagent that was used to probe interactions of untargeted ribosome-nascent chain complexes (11). After cross-linking, samples were extracted with alkaline sodium carbonate buffer to separate integral membrane from peripheral and soluble cross-linked complexes. Immunoprecipitation was used to identify components cross-linked to procoat nascent chains. Neither the translocase subunits SecA and SecY (Fig. 5B, lanes 1 and 2) nor Ffh, the proteinaceous component of the E. coli SRP (Fig. 5B, lane 4), were detected in the supernatant and pellet of the carbonate extraction. The only cross-linking partner that could be identified in this way was the ribosome-associated component trigger factor (Fig. 5B, lane 3), which has been found to interact with all nascent polypeptide chains long enough to protrude from the ribosome (11, 12, 42, 43).
Figure 5.
H1-procoat assembles into the inner membrane by a different mechanism than wild-type procoat. (A) Ribosome–procoat and ribosome–H1-procoat nascent chain complexes, not treated and treated with puromycin, were incubated with inner membrane vesicles and subjected to floatation gradient analysis. The percentage membrane-associated ribosome nascent chain complexes (RNC) was determined. (B) Procoat and H1-procoat ribosome–nascent chain complexes were incubated with inner membrane vesicles and treated with the cross-linker disuccinimidyl suberate. After quenching, soluble and peripheral cross-linking complexes were extracted from the membranes with Na2CO3. Both pellet (pel.) and supernatant (sup.) fractions from trichloroacetic acid-precipitated samples were examined by immunoprecipitation for the presence of cross-linking adducts with the indicated proteins. Immunoprecipitated protein complexes are indicated by an arrow. Relative molecular masses (Mr) of marker proteins are indicated to the right. (C) H1-procoat was expressed in strains CM124 and FF283 not depleted (lane 1) or depleted (lane 2) for SecE or SRP 4.5S RNA, respectively. Cells were pulse-labeled for 15 sec and subsequently processed.
Replacing the Core of the Signal Sequence of M13 Procoat Protein with a Segment from the H1 Transmembrane Helix in Lep Results in SRP- and Preprotein Translocase-Dependent Assembly.
It has been shown that there is a strong correlation between the hydrophobicity of a signal sequence and its affinity for SRP in prokaryotic and eukaryotic systems (10, 11, 42, 44–48). Consistent with this, the three-dimensional structure of the M domain from Thermus aquaticus Ffh includes a large hydrophobic crevice that presumably binds sufficiently hydrophobic signal sequences (48). By using a series of Leu-Ala-based signal sequences of increasing hydrophobicity, a threshold for Ffh binding and efficient secretion of PhoA has been defined at an overall hydrophobicity corresponding to a stretch composed of six Ala and four Leu residues (11, 35, 42). The mean hydrophobicity of the procoat signal peptide was found to be right at the threshold (〈H〉procoat = 2.0–2.1, vs. 〈H〉6A4L = 2.1), suggesting that the lack of Ffh cross-linking and the efficient membrane assembly of procoat in SRP- and SecE-depleted cells might be explained by the low hydrophobicity of the signal sequence. To test whether procoat can be funnelled into the SRP-targeting pathway, the core of the procoat signal sequence was thus replaced with a more hydrophobic stretch from the H1 transmembrane helix in Lep, yielding H1-procoat. The mean hydrophobicity of this construct is higher even than the most hydrophobic PhoA signal peptide tested in the study cited above (〈H〉H1-procoat = 2.7, vs. 〈H〉1A9L = 2.6).
H1-procoat was subjected to the same set of in vitro and in vivo experiments as described above for procoat. H1-procoat nascent chains used in the in vitro studies contained the completed H1-procoat protein. In marked contrast to procoat, floatation gradient analysis showed that H1-procoat ribosome-bound nascent chains associated efficiently with inner membranes (Fig. 5A). This association was enhanced upon release of the nascent chains from the ribosome. Ribosome-nascent chain complexes were also subjected to cross-linking. Similar to procoat, H1-procoat nascent chains could be cross-linked to trigger factor (Fig. 5B, lane 7). In contrast to procoat, however, H1-procoat nascent chains could also be cross-linked to the SRP constituent Ffh in the cytosol (Fig. 5B, lane 8) and to the preprotein translocase component SecA in the membrane (Fig. 5B, lane 5). These observations were consistent with in vivo studies showing that the assembly of H1-procoat was strongly affected under SRP- and SecE-depletion conditions (Fig. 5C). Although it is possible that the effect on processing of H1-procoat is indirectly caused by a lower level of active Lep in cells depleted for SRP or SecE, we see this as unlikely because processing of the wild-type procoat protein and pro-OpmA is not affected under these conditions. Moreover, the level of Lep in depleted cells is the same as in nondepleted cells, as assayed by Western blotting (data not shown).
Lep, Lep-inv, Procoat, and H1-Procoat Do Not Depend on SecB for Efficient Insertion into the Inner Membrane.
Because it has been suggested that SRP dependence correlates with a lack of SecB dependence and vice versa (10, 12), the assembly of Lep, Lep-inv, procoat, and H1-procoat was studied in the E. coli SecB null mutant MM152. Consistent with the suggestion, the assembly of Lep, Lep-inv, or H1-procoat was not affected, whereas procoat was also rapidly converted to mature coat protein in this strain (results not shown). We conclude that SecB is not necessary for the assembly of any of the inner membrane proteins tested herein.
DISCUSSION
We have recently shown that two inner membrane proteins, Lep and Lep-inv, depend on the E. coli SRP for efficient membrane assembly (7). We now show by using a tight SecE-depletion strain that the assembly of these two proteins also depends on the preprotein translocase. Lep has long been known to depend on SecA and SecY for proper assembly, whereas Lep-inv, as well as many other inner membrane proteins with short periplasmic loops, has hitherto been thought not to require a functional preprotein translocase for membrane insertion (21, 49–51). The secA and secY mutant strains used in the earlier studies were primarily selected for defects in protein secretion rather than inner membrane protein assembly, and it is conceivable that in these mutant strains the ability of the preprotein translocase to efficiently export long periplasmic domains is more strongly affected than its ability to assemble short periplasmic loops and transmembrane segments (52).
Because in vivo depletion experiments may suffer from secondary effects, an in vitro cross-linking approach was also used. The in vitro and in vivo results reported herein agree with each other and indicate that the M13 procoat protein does not depend on the SRP-targeting pathway, the SecB chaperone, or the preprotein translocase for proper assembly. Thus inner membrane proteins can bypass the SRP, an observation that is in keeping with the observation that the overexpression of different inner membrane proteins in a strain with artificially depressed SRP levels have differential effects on cell viability, and thus different inner membrane proteins may have different affinities for the SRP (9).
It has been shown that there is a correlation between the hydrophobicity of a signal sequence and its affinity for SRP (10, 11, 42, 44–48). By using a modified procoat construct (H1-procoat) where the hydrophobicity of the signal sequence has been increased, we now demonstrate that an originally SRP- and Sec-independent inner membrane protein can be rerouted into the SRP-dependent pathway, further stressing the importance of signal sequence hydrophobicity. We also note that SRP and preprotein translocase dependence coincide for all proteins tested thus far, strongly supporting the notion that the SRP-targeting pathway is connected to the preprotein translocase in the membrane (12).
In vitro studies using dog pancreas microsomes have shown that Lep and Lep-inv require both the SRP and the ER translocon (Sec61p complex) for proper membrane integration (4), whereas procoat can assemble correctly into the ER membrane in an SRP-independent fashion and appears not to require any other membrane-associated components (22, 53, 54). The basic mechanisms of membrane protein assembly thus seem to be strongly conserved from prokaryotes to eukaryotes.
Acknowledgments
We thank Dr. Chris Murphy for kindly providing strain CM124. This work was supported by a Training and Mobility of Researchers fellowship from the European Commission to J.W.d.G., and by grants from the Swedish Natural Sciences Research Council, the Swedish Cancer Foundation, and the Göran Gustafsson Foundation to G.v.H. Q.V. was supported by the Netherlands Organization for Scientific Research and A.K. was supported by the Deutsche Forschungsgemeinschaft (Grant Ku-749/3). J.L. is the recipient of a Training and Mobility of Researchers project grant from the European Commission.
ABBREVIATIONS
- ER
endoplasmic reticulum
- Ffh
54 homologue
- SRP
signal recognition particle
- Lep
leader peptidase
- IPTG
isopropyl β-d-thiogalactopyranoside
- OmpA
outer membrane protein A
Footnotes
A Commentary on this article begins on page 14587.
References
- 1.Rapoport T A, Jungnickel B, Kutay U. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- 2.High S, Laird V. Trends Cell Biol. 1997;7:206–210. [Google Scholar]
- 3.Johnson A E. Trends Cell Biol. 1997;7:90–95. doi: 10.1016/S0962-8924(97)01029-5. [DOI] [PubMed] [Google Scholar]
- 4.Mothes W, Heinrich S, Graf R, Nilsson I, von Heijne G, Brunner J, Rapoport T. Cell. 1997;89:523–533. doi: 10.1016/s0092-8674(00)80234-2. [DOI] [PubMed] [Google Scholar]
- 5.Hartmann E, Sommer T, Prehn S, Görlich D, Jentsch S, Rapoport T A. Nature (London) 1994;367:654–657. doi: 10.1038/367654a0. [DOI] [PubMed] [Google Scholar]
- 6.MacFarlane J, Müller M. Eur J Biochem. 1995;233:766–771. doi: 10.1111/j.1432-1033.1995.766_3.x. [DOI] [PubMed] [Google Scholar]
- 7.de Gier J-W L, Mansournia P, Valent Q, Phillips G J, Luirink J, von Heijne G. FEBS Lett. 1996;399:307–309. doi: 10.1016/s0014-5793(96)01354-3. [DOI] [PubMed] [Google Scholar]
- 8.Seluanov A, Bibi E. J Biol Chem. 1997;272:2053–2055. doi: 10.1074/jbc.272.4.2053. [DOI] [PubMed] [Google Scholar]
- 9.Ulbrandt N D, Newitt J A, Bernstein H D. Cell. 1997;88:187–196. doi: 10.1016/s0092-8674(00)81839-5. [DOI] [PubMed] [Google Scholar]
- 10.de Gier J-W L, Valent Q, von Heijne G, Luirink J. FEBS Lett. 1997;408:1–4. doi: 10.1016/s0014-5793(97)00402-x. [DOI] [PubMed] [Google Scholar]
- 11.Valent Q A, de Gier J W L, von Heijne G, Kendall D A, ten Hagen-Jongman C M, Oudega B, Luirink J. Mol Microbiol. 1997;25:53–64. doi: 10.1046/j.1365-2958.1997.4431808.x. [DOI] [PubMed] [Google Scholar]
- 12.Valent Q A, Scotti P A, High S, de Gier J W L, von Heijne G, Lentzen G, Wintermeyer G, Oudega B, Luirink J. EMBO J. 1998;17:2504–2512. doi: 10.1093/emboj/17.9.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersson H, von Heijne G. EMBO J. 1993;12:683–691. doi: 10.1002/j.1460-2075.1993.tb05702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Heijne G. FEBS Lett. 1994;346:69–72. doi: 10.1016/0014-5793(94)00296-7. [DOI] [PubMed] [Google Scholar]
- 15.von Heijne G. Membrane Protein Assembly. Austin, TX: Landes; 1997. pp. 55–62. [Google Scholar]
- 16.von Heijne G, Manoil C. Protein Eng. 1990;4:109–112. doi: 10.1093/protein/4.2.109. [DOI] [PubMed] [Google Scholar]
- 17.Nilsson I M, von Heijne G. Cell. 1990;62:1135–1141. doi: 10.1016/0092-8674(90)90390-z. [DOI] [PubMed] [Google Scholar]
- 18.Kuhn A, Kreil G, Wickner W. EMBO J. 1986;5:3681–5. doi: 10.1002/j.1460-2075.1986.tb04699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolfe P B, Rice M, Wickner W. J Biol Chem. 1985;260:1836–1841. [PubMed] [Google Scholar]
- 20.Lee J I, Kuhn A, Dalbey R E. J Biol Chem. 1992;267:938–943. [PubMed] [Google Scholar]
- 21.von Heijne G. Nature (London) 1989;341:456–458. doi: 10.1038/341456a0. [DOI] [PubMed] [Google Scholar]
- 22.Wickner W. Biochemistry. 1988;27:1081–1086. doi: 10.1021/bi00404a001. [DOI] [PubMed] [Google Scholar]
- 23.Kuhn A. Eur J Biochem. 1988;177:267–271. doi: 10.1111/j.1432-1033.1988.tb14372.x. [DOI] [PubMed] [Google Scholar]
- 24.Wickner W, Leonard M R. J Biol Chem. 1996;271:29514–29516. doi: 10.1074/jbc.271.47.29514. [DOI] [PubMed] [Google Scholar]
- 25.Yang Y B, Yu N J, Tai P C. J Biol Chem. 1997;272:13660–13665. doi: 10.1074/jbc.272.21.13660. [DOI] [PubMed] [Google Scholar]
- 26.Randall L, T B, T, Hardy S J S, Pavlov M Y, Freistroffer D V, Ehrenberg M. Proc Natl Acad Sci USA. 1997;94:802–805. doi: 10.1073/pnas.94.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ribes V, Römisch K, Giner A, Dobberstein B, Tollervey D. Cell. 1990;63:591–600. doi: 10.1016/0092-8674(90)90454-m. [DOI] [PubMed] [Google Scholar]
- 28.Kumamoto C A, Beckwith J. J Bacteriol. 1985;163:267–274. doi: 10.1128/jb.163.1.267-274.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neidhardt F C, Bloch P L, Smith D F. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Traxler B, Murphy C. J Biol Chem. 1996;271:12394–12400. doi: 10.1074/jbc.271.21.12394. [DOI] [PubMed] [Google Scholar]
- 31.Johnston S, Lee J H, Ray D S. Gene. 1985;34:137–145. doi: 10.1016/0378-1119(85)90121-0. [DOI] [PubMed] [Google Scholar]
- 32.Fürste J P, Pansegrau W, Frank R, Blöcker H, Scholz P, Bagdasarian M, Lanka E. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 33.Schägger H, von Jagow F. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Doud S K, Chou M M, Kendall D A. Biochemistry. 1993;32:1251–1256. doi: 10.1021/bi00056a008. [DOI] [PubMed] [Google Scholar]
- 36.Claros M G, von Heijne G. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 37.Engelman D M, Steitz T A, Goldman A. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- 38.Segrest J P, De Loof H, Dohlman J G, Brouilette C G, Anantharamaiah G M. Proteins. 1990;8:103–117. doi: 10.1002/prot.340080202. [DOI] [PubMed] [Google Scholar]
- 39.Gallusser A, Kuhn A. EMBO J. 1990;9:2723–2729. doi: 10.1002/j.1460-2075.1990.tb07459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soekarjo M, Eisenhawer M, Kuhn A, Vogel H. Biochemistry. 1996;35:1232–1241. doi: 10.1021/bi951087h. [DOI] [PubMed] [Google Scholar]
- 41.Akiyama Y, Kihara A, Tokuda H, Ito K. J Biol Chem. 1996;271:31196–31201. doi: 10.1074/jbc.271.49.31196. [DOI] [PubMed] [Google Scholar]
- 42.Valent Q A, Kendall D A, High S, Kusters R, Oudega B, Luirink J. EMBO J. 1995;14:5494–5505. doi: 10.1002/j.1460-2075.1995.tb00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hesterkamp T, Bukau B. FEBS Lett. 1996;389:32–34. doi: 10.1016/0014-5793(96)00582-0. [DOI] [PubMed] [Google Scholar]
- 44.High S, Henry R, Mould R M, Valent Q, Meacock S, Cline K, Gray J C, Luirink J. J Biol Chem. 1997;272:11622–11628. doi: 10.1074/jbc.272.17.11622. [DOI] [PubMed] [Google Scholar]
- 45.Ng D T W, Brown J D, Walter P. J Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hatsuzawa K, Tagaya M, Mizushima S. J Biochem (Tokyo) 1997;121:270–277. doi: 10.1093/oxfordjournals.jbchem.a021583. [DOI] [PubMed] [Google Scholar]
- 47.Matoba S, Ogrydziak J Biol Chem. 1998;273:18841–18847. doi: 10.1074/jbc.273.30.18841. [DOI] [PubMed] [Google Scholar]
- 48.Keenan R J, Freymann D M, Walter P, Stroud R M. Cell. 1998;94:181–191. doi: 10.1016/s0092-8674(00)81418-x. [DOI] [PubMed] [Google Scholar]
- 49.Bassilana M, Gwizdek C. EMBO J. 1996;15:5202–5208. [PMC free article] [PubMed] [Google Scholar]
- 50.Werner P K, Saier M H, Müller M. J Biol Chem. 1992;267:24523–24532. [PubMed] [Google Scholar]
- 51.Yamato I. J Biochem (Tokyo) 1992;111:444–450. doi: 10.1093/oxfordjournals.jbchem.a123777. [DOI] [PubMed] [Google Scholar]
- 52.Duong F, Wickner W. EMBO J. 1998;17:696–705. doi: 10.1093/emboj/17.3.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watts C, Wickner W, Zimmermann R. Proc Natl Acad Sci USA. 1983;80:2809–2813. doi: 10.1073/pnas.80.10.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dierks T, Volkmer J, Schlenstedt G, Jung C, Sandholzer U, Zachmann K, Schlotterhose P, Neifer K, Schmidt B, Zimmermann R. EMBO J. 1996;15:6931–6942. [PMC free article] [PubMed] [Google Scholar]