Abstract
The parasitic bacterium Mycoplasma genitalium has a small, reduced genome with close to a basic set of genes. As a first step toward determining the families of protein domains that form the products of these genes, we have used the multiple sequence programs psi-blast and geanfammer to match the sequences of the 467 gene products of M. genitalium to the sequences of the domains that form proteins of known structure [Protein Data Bank (PDB) sequences]. PDB sequences (274) match all of 106 M. genitalium sequences and some parts of another 85; thus, 41% of its total sequences are matched in all or part. The evolutionary relationships of the PDB domains that match M. genitalium are described in the structural classification of proteins (SCOP) database. Using this information, we show that the domains in the matched M. genitalium sequences come from 114 superfamilies and that 58% of them have arisen by gene duplication. This level of duplication is more than twice that found by using pairwise sequence comparisons. The PDB domain matches also describe the domain structure of the matched sequences: just over a quarter contain one domain and the rest have combinations of two or more domains.
The structural, functional, and evolutionary unit in proteins is called a domain. Small proteins, and many medium-sized proteins, contain just one domain; larger proteins are formed by combinations of domains (1, 2). The evolution of development has involved gene duplication, divergence, and, in many cases, domain rearrangements (3–7). With the advent of completely sequenced genomes, we can begin to define the repertoire of domains, and the duplications and combinations, that have produced the proteins that occur in different organisms (6–12). This will help us to understand the molecular basis of the different properties of organisms and their evolution.
Ideally, the repertoire of domains and their arrangements in proteins would be found by direct comparisons of these sequences to each other. However, protein sequences can diverge to such an extent that these types of comparisons fail to detect evolutionary relationships (13). For related proteins that have sequence identities of 20–30%, only one-half of the relationships can be detected by pairwise sequence comparisons, and for related proteins with lower identities, the proportion is much smaller (14). This means that, if a high proportion of related proteins in organisms have identities of <30%, pairwise comparisons will detect only a small fraction of the domain duplications and rearrangements that produced them.
There are two ways of overcoming, at least in part, the limitations of pairwise sequence comparisons. First, the current comparison methods that use multiple sequences, while still failing to detect many distant relationships, are three times as effective as pairwise comparisons (15). Second, on a different level, if the structures of the proteins being compared are known, distant evolutionary relationships can usually be detected from the combination of sequence, structural, and functional information (1, 16). This means that if sequences from the genome can be matched to sequences of proteins of known structure, we can usually determine whether they are related.
In this paper, we use the multiple sequence comparison program psi-blast (17) to match the sequences of the domains of proteins of known structure (PDBD sequences) to the sequences produced by the genome of Mycoplasma genitalium (MG) (18). This parasitic bacterium has a reduced simplified genome with 467 genes for potential proteins. The number of significant sequence matches made by psi-blast is twice that made by pairwise comparisons: 191 MG sequences are matched in all or in part by the sequences of 274 PDBD domains. Altogether these matches cover over a quarter (27%) of the MG genome in terms of amino acid residues. The evolutionary relationships of the PDB domains that match the MG sequences show that the extent of domain duplication in MG is at least twice that suggested by pairwise comparisons. They also show extensive rearrangements and combinations of domains.
Sequences and Sequence Families in MG.
Translations of the MG ORFs were obtained from The Institute for Genomic Research through its web address http://www.tigr.org/. MG has 467 genes, which potentially code for proteins. The MG sequences are annotated with either an indication of their function, if this is known directly or if the sequence is significantly similar to that of a protein known function, or as a “hypothetical protein,” if the sequence is similar to a that of a protein of unknown function. In the current MG database, 316 sequences have some functional annotation and 151 are hypothetical proteins. Of the 151, there are 96 that are homologous only to proteins in the M. pneumoniae genome (19).
To determine relationships between MG sequences that can be found by pairwise sequence comparisons we used the genome analysis and protein family maker (geanfammer) procedure (20). The main steps in this procedure are (i) an all-against-all comparison of the sequences by using fasta ktup = 1 (21); (ii) single-linkage grouping of all genes found to be similar beneath the threshold expectation (E) value of 0.01 into the same family, and (iii) resolution of complex families that have indirect matches between members. An example of a complex family would be given by three proteins 1, 2, and 3 built, respectively, from domains A and B, B and C, and C and D. Proteins 1 and 3 are not related, but simple single-linkage clustering will put all three in the same family because the different parts of 2 match 1 and 3. The divclus program in geanfammer resolves these complex multidomain families (a full description is given in ref. 20).
The initial grouping produced 46 families. Resolution of the complex families increased this to 84. These range in size from 59 families each with two members, to one family with 14 members; see Table 1 for details. Ninety-three MG proteins belong to only one family, 20 have domains belonging to two different families, and 20 have domains belonging to between 3 and 9 different families.
Table 1.
Protein families formed by the pairwise comparisons of MG sequences and the resolution of complex families by geanfammer
Whole sequences.
Unmatched regions of matched sequences.
n, number of sequences in family.
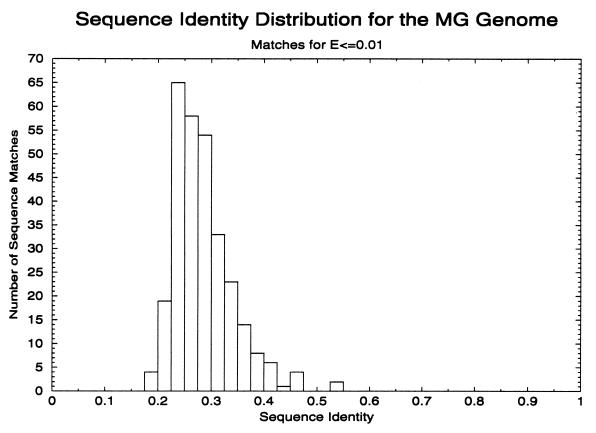
It should be emphasized here that the limited ability of pairwise sequence comparison methods to find distant relatives means that these calculations underestimate the number of related genes. In Fig. 1 we show the residue identities of the pairs of MG sequence that form these families. Only 91 pairs have residue identities >30%, so most of the matches come from regions where the ability of pairwise comparisons to detect relationships is very limited (14). The match of PDB sequences to structures, described below, does indeed show that pairwise sequence comparisons detect only a small fraction of relationships actually present in the MG sequences. (Use of a higher E value criteria for true relationships will give a number of additional families and increase the size of others but at the cost of a higher error rate.)
Figure 1.
Sequence identity histogram of all protein pairs in the MG genome that match at an E value of ≤0.01 by using fasta ktup = 1. Self matches are excluded. Most pairs of proteins match each other in the region below 30%, which is the region where the ability to detect relationships by pairwise sequence comparison programs drops off rapidly.
Sequences of Domains in Proteins of Known Structure and Their Evolutionary Relationships.
The Structural Classification of Proteins (SCOP) database contains a description of the evolutionary and structural relations of those proteins whose atomic structure has been determined (1). The current version is available on the world wide web (WWW) at http://scop.mrc-lmb.cam.ac.uk/scop/.
The unit of classification in the database is the structural, functional, and evolutionary unit of proteins: the domain. The sequences corresponding to these domains will be denoted as PDBD sequences. As was mentioned above, small proteins and most of those of medium size have a single domain and are, therefore, treated as a whole. The domains that form large proteins are classified individually, if there is evidence from different protein structures that they are evolutionary units that can undergo independent duplication and recombination. (Large proteins that contain domains that, up to now, are seen linked only to each other are treated as a single unit in SCOP: these entries form 9% of the superfamily entries in the current version of the database. It is possible that these domains are effective only when they are linked to each other and that they do not undergo recombination separately. However, we do expect that, in most cases, future structures will provide evidence for their being separate evolutionary units.)
Domains are clustered together into families if they have close evolutionary relationships. Superfamilies bring together families whose proteins have low sequence identities but whose structural details and, in many cases, functional features suggest that a common evolutionary origin is very probable; for example, the variable and constant domains of immuno-globulins. The fold classification brings together superfamilies that have the same secondary structures in the same arrangement. For different superfamilies that share a common fold, there is usually no evidence that they have evolutionary relationships in most cases. In a few cases, the situation is less clear because there is weak evidence that does suggest the existence of evolutionary relationships. In these less certain cases, the superfamilies are kept separate until the subsequent discovery of intermediate structures that provide stronger support for their merger (15).
Many of the ≈7,000 structures in the current PDB (22) are variants of a smaller number of native proteins that differ, for example, only in the presence or absence of a substrate or an engineered mutation. For the purposes of this work, it is useful to remove this redundancy and work with a set of sequences that have residue identities of no greater than 95% to one another: PDB95D. Such a set is available from the current release of the SCOP database (version 1.37, February 1998) at http://scop.mrc-lmb.cam.ac.uk/scop/pdbd.html and will be brought up to date in subsequent SCOP releases.
In the work described here, we used an edited version of PDB95D from which we removed those sequence that have characteristics that are likely to produce false matches with low E values. These include proteins with high cysteine content and leucine-rich repeats. This version of PDB95D, called PDB95D-T, has 2,231 sequences. It is available from the SCOP URL given above. The 2,231 sequences belong to one of 509 superfamilies. Of these superfamilies, 230 are represented by a single sequence; 225 by 2–9 sequences, and 64 by 10–312 sequences.
psi-blast Match of PDB95D-T Sequences to MG Sequences.
The psi-blast program (17) was used to match the PDB95D-T sequences and MG sequences. psi-blast begins with an initial gapped blast search that collects, from a sequence database, homologs that match a query sequence with E values below a threshold set by the user. A position-specific matrix is built from an alignment of these sequences. The sequence database is then searched with this profile, and sequences that match with a score below the threshold are used to build a new matrix for the next round of searching. This process is continued either to convergence, i.e., to the point in which no new sequences are found by the profile, or until the iteration number specified by the user is reached. Here we used the psi-blast parameters that had been found to be effective in giving a good coverage to error ratio: an E value threshold of 0.0005 for the selection of homologous sequences for the psi-blast profile; allowing up to 20 iterative searches and an E value threshold of 10−5 to be considered significant for match between the query profile and a target sequence (15). The error per query using these values was estimated to be ≈1%.
Two psi-blast searches were carried out. In the first, PDB95D-T sequences were used to query MG sequences embedded in the nonredundant sequence database (NRDB) NRDB90, (February 1998 version) (23). The second search was the reverse of the first: MG sequences were used as queries to search for the PDB95D-T sequences embedded in the NRDB90. These two searches were carried out because pairs of homologous query sequences will find different close homologs at the first stage of a psi-blast search and hence build different profiles. This means there are some cases where sequence A can find a match to B, but not B to A.
In the searches, we used two criteria for a significant match of a profile of a PDB95-T sequence to an MG sequence: (i) an E value score of ≤10−5 (see above) and (ii) the matched region had to be at least one-half the length of the PDB95D-T sequence and not less than 30 residues.
There are many cases in which one or more PDB95D-T sequences matched different regions of an MG sequence, and these matches are examples of domain combinations. However, there were also seven cases where PDB95D-T sequences belonging to different superfamilies had matched the same region of an MG sequence with similar good scores. These were discarded.
A number of MG sequences are not matched by a PDB95D-T sequences in the psi-blast calculations but are members of geanfammer sequence families, some of whose members are matched. On the intermediate sequence principle, these unmatched sequences should be seen as distant homologs of the PDB95D-T sequence that matches other members of the family. Taking into account a reasonable match region, 5 MG sequences have an indirect match to a PDB95D-T sequence.
The net result of these calculations is that 191 MG sequences are matched all or in part by 274 PDB95D-T sequences. Individual MG sequences are matched in nonoverlapping regions by between one and six PDB95D-T sequences, and a general view of the number of domains found in the MG sequences is given in Table 1. In the discussion of these results, it is useful to give the MG sequences and sequence regions matched by PDB sequences a specific name. We will refer to these as MGPDB sequences and MGPDB sequence regions; respectively. These 191 MGPDB sequences are 41% of the 467 ORFs in MG. Full details of the matches are available from http://www.mrc-lmb.cam.ac.uk/genomes/MG_strucs.html.
For a quarter of the sequences whose function was previously unknown, the psi-blast/geanfammer results provide both structural and functional information. In the case of MG, 37 sequences with a “hypothetical protein” annotation are matched to a structure. This number reduces the fraction of proteins without any informative annotation from 32% to 24% of the MG genome. The sequence families found by geanfammer and not assigned a structure are potential targets for “structural genomics” projects (24, 25), which have as their aim the solution of one protein structure in each family.
Previous Matches of PDB and MG Sequences.
Previous attempts to match PDB sequences to MG sequences by using pairwise comparisons have usually produced less than one-half the number of matches described here. For the early attempts, this was partly because fewer PDBD sequences were known. However, we find that matching the present PDB95D-T sequence to the MG sequences by using one of the most effective pairwise sequence comparison methods, fasta ktup = 1 (21) calibrated for a 1% error rate (14), assigns structures to 19% of the proteins in MG. This implies that the use of multiple sequences in psi-blast procedure is major reason for finding twice as many matches.
Fischer and Eisenberg (26) used the Smith–Waterman pairwise sequence comparison algorithm and a fold recognition server to assign structures to 22% of the proteins in MG. Of the 22%, 16% were assigned by pairwise comparisons. The 6% found by a fold recognition method consists of 28 proteins. The psi-blast procedure described here makes the same structural assignment for 22 of the 28 sequences and different assignments for two sequences. A structure that is clearly homologous to one of these proteins (MG367) has been solved (27) and shows that the assignment suggested here (a double-stranded RNA-binding domain) is correct. In addition, the fold recognition assignment to one of the four sequences not found with our psi-blast method (MG088) has been shown to be incorrect because the structure of a homologous protein has since been solved (28).
After the completion of the calculations described here, Huynen et al. (29) reported that by using psi-blast they find all or part of 35% of MG sequences have significant matches to PDB domains. The main reason for the somewhat smaller number of matches is that they carried out only one search; not the two used here (see above).
Superfamilies of Domains MGPDB Sequences.
As mentioned above, we call the MG sequences and sequence regions that are matched by PDB sequences MGPDB sequences and MGPDB sequence regions. The superfamily assignments of the 274 MGPDB sequence regions are derived from the SCOP superfamily of the matching PDB sequence. Of the 274 sequences, 72 are the only representatives of their superfamilies. The other 202 belong to one of 43 superfamilies that have between 2 and 51 members; see Table 2. Thus, the 274 MGPDB sequences belong to one of 72 + 43 = 115 superfamilies.
Table 2.
Number and size of families formed by the MGPDB sequences
| Family size, n | psi-blast and SCOP superfamilies | fasta, ktup = 1 sequence families |
|---|---|---|
| 1 | 72 | 178 |
| 2 | 22 | 24 |
| 3 | 7 | 4 |
| 4 | 6 | — |
| 5 | 2 | — |
| 8 | 1 | — |
| 9 | 1 | 2 |
| 10 | 1 | — |
| 12 | 1 | — |
| 13 | 1 | — |
| 18 | — | 1 |
| 51 | 1 | — |
| Total sequences | 274 | 274 |
n, number of sequences in a family.
The functions of the eight largest superfamilies are described in Table 3. They have between 5 and 51 members and contain 42% of the MGPDB sequences regions. Fifty percent of the MGPDB sequences have one domain, or occasionally two, that are a member of one, or two, of these eight superfamilies. Descriptions of functions of the smaller families can be found at http://www.mrc-lmb.cam.ac.uk/genomes/MG_fams.html.
Table 3.
Functions of large superfamilies formed by the MGPDB sequences
| No. of MG sequences in superfamily | SCOP description of superfamily |
|---|---|
| 51 | P-loop nucleotide triphosphate hydrolases |
| 13 | Class I aminoacyl-tRNA synthetases, catalytic domain |
| 12 | Three nucleotide-binding domains |
| 10 | Class II aminoacyl-tRNA synthetases |
| 9 | Nucleic acid-binding proteins |
| 8 | Type II DNA topoisomerase |
| 5 | Translation factors |
| 5 | FAD/NAD-linked reductases, dimerisation domain |
The evolutionary relationships based on structure, sequence, and function, and described in SCOP, create superfamilies of MG sequences that are an amalgam of the singlets and sequences families produced by the geanfammer calculations described above. For example, the largest family, the 51 sequences in the P-loop containing nucleotide triphosphate hydrolases, brings together 24 single sequences (of which three sequences contain duplications of this domain); one family with 14 members (ABC transporters), five families with two members, and one family with three members.
The large size in MG of three of the families in Table 3, the P-loop nucleotide triphosphate hydrolases, Rossmann nucleotide-binding domains, and nucleic acid-binding proteins has been also discussed by Koonin et al. (30).
Gerstein (31) matched PDB sequences to sequences from the genomes of Haemophilus influenzae, Methanococcus jannaschii, and Saccharomyces cerevisiae. In all three organisms, he found that three of the four largest families are the P-loop containing nucleotide triphosphate hydrolases, Rossmann nucleotide-binding domains, and α/β (TIM) barrels. In MG, the P-loop containing nucleotide triphosphate hydrolases are the largest family and the Rossmann nucleotide-binding domains are the third largest family. In our work on MG, α/β barrels are put in a number of separate superfamilies, but if these folds are brought together, they make the sixth largest family.
Gene Duplications in MGPDB Sequences.
Two measures have been used to describe the overall extent to which gene duplications have occurred in a genome. One of these is the proportion of all sequences that have arisen by gene duplications. (When we refer to gene duplication, we mean the duplication of the part of a gene that corresponds to one protein domain; or, in the case of SCOP multidomain entries, the part of the gene that corresponds to that particular combination of domains.) For the MGPDB sequences, the number and size of superfamilies is given in Table 2. There are 72 MGPDB sequences that are not related to any other MGPDB sequence, and these can be seen as forming 72 single member families. There are 43 superfamilies that have between 2 and 51 members and, in all, they contain 202 MGPDB sequences. As, by definition, members of the same superfamily are descended from a common ancestor, the creation of these families involved 202 − 43 = 159 gene duplications. Thus, for the MGPDB sequences, the proportion that have arisen by gene duplication is 159/274, i.e., 58%.
The second measure of gene duplications is the proportion of sequences that are in families. For the MGPDB sequences, this is 202/274, i.e., 74%. These figures are much larger than those obtained by the simple pairwise comparisons of MG sequences. Above we described the results of the geanfammer calculations in which family relationships are found by pairwise comparisons (fasta, ktup = 1), and in Table 1, we give the number and size of these families. Calculations by using this data give the proportion of MG sequences that have arisen by gene duplication as 24% and the proportion that are in families as 28%. Using a combination of automatic methods and expertise, pairwise sequence comparisons, visual analysis of multiple alignments, and protein motifs, Koonin et al. (30) calculated that 35% of all MG sequences are members of protein families.
However these numbers, derived from calculations by using all MG sequences, might be thought inappropriate for comparisons with numbers derived from MGPDB sequences that form only 27% of the genome. This would be the case if gene duplications in the matched regions are much more common than those in the unmatched regions. To check this, the MGPDB sequences were collected, compared with each other by using fasta, ktup = 1, and matched pairs with E values ≤0.01 clustered into families (Table 2). The distribution of MGPDB sequences in families that is given by these calculation is very close to that for all MG sequences. In each case, close to two-thirds of the sequences are singlets, a quarter in families with two or three members and one-eighth in larger families (see Tables 1 and 2). Also the proportion of MGPDB sequences that the sequence comparisons give as having arisen by gene duplication, 24%, and the proportion that are in families, 35%, are close to the proportions given for all MG sequences: 24% and 28%. Thus, the extent to which MGPDB sequences are similar to each other is very similar that extent to which all MG sequences are similar to each other.
Our calculations with psi-blast and SCOP give rates of gene duplications that are more than twice greater than those based on sequence comparisons because (i) the psi-blast procedure finds three times as many true matches for distantly related sequences and (ii) the SCOP relationships, based on a combination of sequence, structure and functional information, include many that cannot be found by pairwise comparisons or psi-blast (see Table 2 and ref. 15). Indeed, the extent of gene duplications described here is likely to be an underestimate of the true proportion, because psi-blast is still only partially successful in detecting distant relationships (15), and, therefore, it is very likely that some of the unmatched MG sequences will be distant homologs of some of the matched sequences. Koonin et al. (30), who used multiple alignment analysis and protein motifs in their analysis of MG, found more members for one of the families discussed here: 56 rather than 51 members for the P-loop containing nucleotide triphosphate hydrolases.
Combinations of Domains in MGPDB Sequences.
The match of PDB sequences gives detailed information on the domain structure of all or part of 41% of MG sequences. Table 4 gives the statistics for the number of domains that match MGPDB sequences. There are 106 MG sequences in which the PDBD matches cover the whole sequence. Fifty-nine of these are wholly matched by a single PDBD sequence. Another 40 MG sequences are wholly matched by between 2 and 5 PDB domains.
Table 4.
PDBD domains assigned to MG sequences
| PDB95D-T sequences that match MG sequences, n | MG sequences matched by PDB95D-T sequences, n
|
||
|---|---|---|---|
| Matches to the whole MG sequences | Matches to part of the MG sequences | Totals | |
| 1 | 59 + 7 | 62 + 4 | 121 + 11 |
| 2 | 27 | 11 + 3 | 38 + 3 |
| 3 | 9 | 5 | 14 |
| 4 | 2 | — | 2 |
| 5 | 2 | — | 1 |
| Totals | 99 + 7 = 106 | 78 + 7 = 85 | 177 + 14 = 191 |
Matches can be made to MG sequences by one or more of the many single domain PDB95-T sequences or one of the few multidomain sequences. When the number of matches is given as j or j + k, j is the number of matches made by a single domain PDB95D-T sequences. If n = 1, k is the number of matches made by one multidomain PDB95D-T sequences. If n = 2, k is the number of matches made by one single domain sequence together with one multidomain sequence.
There are also 7 sequences wholly matched by one of the SCOP multidomain entries. (The multidomain entries are for proteins whose domains are only seen linked to each other up to now, see above. We expect that, in most cases, future structures will provide evidence for their domains being separate evolutionary units.)
There are 85 MG sequences that are partly matched by between one and three PDBD sequences and the unmatched region is long enough for at least one additional domain. These results mean that more than two-thirds of the MGPDB sequence are formed by combinations of domains. Another aspect of the combinatorial patterns of domains is the observation that, of the 59 domains that match whole MG sequences, only 23 are restricted to single domain proteins; the other 36 have homologs in the multidomain MG proteins.
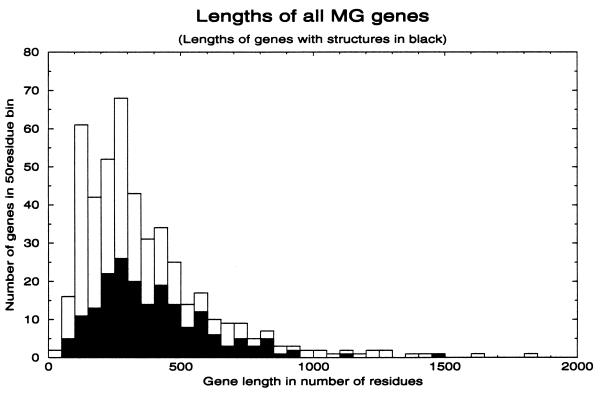
To see whether these general results on the domain structure of the 41% of MG proteins could also apply to some of the other 59% of MG proteins, we check whether the MGPDB sequences have a length distribution skewed in comparison with unmatched MG sequences. The histogram in Fig. 2 shows the length distribution of sequences assigned structures superimposed on the distribution of all sequences in the MG genome. It is obvious that the sequences with structures are distributed across all lengths in a way similar to that of all sequences in the genome. This result indicates that the large majority of proteins in the MG genome have involved rearrangement of domains.
Figure 2.
Histogram of lengths of all protein sequences in MG. Sequences are placed in bins with steps of 50 residues. The lengths of genes that match PDBD sequences are superimposed in solid black. The distribution of matched genes is approximately the same as that of the whole genome.
CONCLUSIONS
Our results demonstrate clearly that structural assignments to genome sequences, and the knowledge that this gives of their evolutionary relationships, are required for the accurate determination of the repertoire of domains, duplications, and recombinations that have formed the genomes in different organisms. The use of psi-blast and geanfammer to match MG sequences to PDB sequences, together with the information available on evolutionary relationships of PDB sequences in SCOP, indicate levels of gene duplication in MG that are about twice greater than those given by pairwise comparisons. Using these procedures on other genomes would also give much higher values than those found by pairwise comparisons. The effectiveness of these kinds of calculations is likely to increase rapidly. X-ray crystallography and NMR are producing many new structures each year. Hidden Markov models of aligned sequences (32, 33) can detect evolutionary relationships not found by psi-blast (15) and should soon be available for easy automatic use.
Acknowledgments
We thank Alex Bateman, Alexey Murzin, and Bissan Al-Lazikani for advice and help with the initial stages of this work and thank Prof. G. Church. We are grateful to Peer Bork for helpful discussions and information before publication. S.A.T. has a predoctoral fellowship from the Boehringer Ingelheim Fonds. J.P. has a grant from Hoechst-Marion Roussel. This paper is dedicated to Walter Henderson.
ABBREVIATIONS
- MG
Mycoplasma genitalium
- PDBD
domains of proteins of known structure
- E value
expectation value
- SCOP
structural classification of proteins database
- PDB95D-T
library of sequences of domains of proteins with known structure filtered at 95% sequence identity
Note Added in Proof
Using new PDB sequences, we now find that 301 domains match all or part of 210 genes and cover 31% of the MG genome. Details are given at http://www.mrc-lmb.cam.ac.uk/genomes/MG_strucs.html.
References
- 1.Murzin A G, Brenner S E, Hubbard T, Chothia C. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 2.Orengo C A, Michie A D, Jones S, Jones D T, Swindells M B, Thornton J M. Structure (London) 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 3.Ingram V. Nature (London) 1961;189:704–708. doi: 10.1038/189704a0. [DOI] [PubMed] [Google Scholar]
- 4.Rossmann M G, Moras D, Olsen K W. Nature (London) 1974;250:194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- 5.Patthy L. Curr Opin Struct Biol. 1991;1:351–361. [Google Scholar]
- 6.Riley M, Labedan B. J Mol Biol. 1997;268:857–868. doi: 10.1006/jmbi.1997.1003. [DOI] [PubMed] [Google Scholar]
- 7.Henikoff S, Greene E A, Pietrokovski S, Bork P, Attwood T K, Hood L. Science. 1997;278:609–614. doi: 10.1126/science.278.5338.609. [DOI] [PubMed] [Google Scholar]
- 8.Chothia C. Nature (London) 1992;357:543–544. doi: 10.1038/357543a0. [DOI] [PubMed] [Google Scholar]
- 9.Green P, Lipman D, Hillier L, Waterson R, States D, Claverie J M. Science. 1993;259:1711–1716. doi: 10.1126/science.8456298. [DOI] [PubMed] [Google Scholar]
- 10.Brenner SE, Hubbard T, Murzin A, Chothia C. Nature (London) 1995;378:140. doi: 10.1038/378140a0. [DOI] [PubMed] [Google Scholar]
- 11.Gerstein M, Levitt M. Proc Natl Acad Sci USA. 1997;94:11911–11916. doi: 10.1073/pnas.94.22.11911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tatusov R L, Koonin E V, Lipman D J. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 13.Doolittle R F. Of URFs and ORFs: A Primer on How to Analyze Derived Amino Acid Sequences. Mill Valley, CA: University Science Books; 1987. [Google Scholar]
- 14.Brenner S E, Chothia C, Hubbard T J P. Proc Natl Acad Sci USA. 1998;95:6073–6078. doi: 10.1073/pnas.95.11.6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park J, Karplus K, Barrett C, Hughey R, Haussler D, Hubbard T, Chothia C. J Mol Biol. 1998;284:1201–1210. doi: 10.1006/jmbi.1998.2221. [DOI] [PubMed] [Google Scholar]
- 16.Brenner S E, Chothia C, Hubbard T J P, Murzin A G. Methods Enzymol. 1996;266:635–643. doi: 10.1016/s0076-6879(96)66039-x. [DOI] [PubMed] [Google Scholar]
- 17.Altschul S F, Madden T L, Schaffer A A, Zhang J H, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, et al. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 19.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B-C, Herrmann R. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park J, Teichmann S A. Bioinformatics. 1998;14:144–150. doi: 10.1093/bioinformatics/14.2.144. [DOI] [PubMed] [Google Scholar]
- 21.Pearson W R, Lipman D J. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernstein F C, Koetzle T F, Williams G J B, Meyer E F, Brice M D, Rodgers J R, Kennard O, Shimanouchi T, Tasumi M. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 23.Holm L, Sander C. Bioinformatics. 1998;14:423–429. doi: 10.1093/bioinformatics/14.5.423. [DOI] [PubMed] [Google Scholar]
- 24.Rost B. Structure (London) 1998;6:259–263. doi: 10.1016/s0969-2126(98)00029-x. [DOI] [PubMed] [Google Scholar]
- 25.Shapiro L, Lima C D. Structure (London) 1998;6:265–267. doi: 10.1016/s0969-2126(98)00030-6. [DOI] [PubMed] [Google Scholar]
- 26.Fischer D, Eisenberg D. Proc Natl Acad Sci USA. 1997;94:11929–11934. doi: 10.1073/pnas.94.22.11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kharat A, Nacias M J, Gibson T J, Nilges M, Pastore A. EMBO J. 1995;14:3572–3584. doi: 10.1002/j.1460-2075.1995.tb07363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wimberly B T, White S W, Ramakrishnan V. Structure (London) 1997;5:1187–1198. doi: 10.1016/s0969-2126(97)00269-4. [DOI] [PubMed] [Google Scholar]
- 29.Huynen M, Doerks T, Eisenhaber F, Orengo C, Sunyaev S, Yuan Y, Bork P. J Mol Biol. 1998;280:323–326. doi: 10.1006/jmbi.1998.1884. [DOI] [PubMed] [Google Scholar]
- 30.Koonin E V, Mushegian A R, Galperin M Y, Walker D R. Mol Microbiol. 1997;25:619–637. doi: 10.1046/j.1365-2958.1997.4821861.x. [DOI] [PubMed] [Google Scholar]
- 31.Gerstein M. J Mol Biol. 1997;274:562–576. doi: 10.1006/jmbi.1997.1412. [DOI] [PubMed] [Google Scholar]
- 32.Eddy S R. Curr Opin Struct Biol. 1996;6:361–365. doi: 10.1016/s0959-440x(96)80056-x. [DOI] [PubMed] [Google Scholar]
- 33.Krogh A, Brown M, Mian I S, Sjolander K, Haussler D. J Mol Biol. 1994;235:1501–1531. doi: 10.1006/jmbi.1994.1104. [DOI] [PubMed] [Google Scholar]