Abstract
A multiple protein–DNA complex formed at a human α-globin locus-specific regulatory element, HS-40, confers appropriate developmental expression pattern on human embryonic ζ-globin promoter activity in humans and transgenic mice. We show here that introduction of a 1-bp mutation in an NF-E2/AP1 sequence motif converts HS-40 into an erythroid-specific locus-control region. Cis-linkage with this locus-control region, in contrast to the wild-type HS-40, allows erythroid lineage-specific derepression of the silenced human ζ-globin promoter in fetal and adult transgenic mice. Furthermore, ζ-globin promoter activities in adult mice increase in proportion to the number of integrated DNA fragments even at 19 copies/genome. The mutant HS-40 in conjunction with human ζ-globin promoter thus can be used to direct position-independent and copy number-dependent expression of transgenes in adult erythroid cells. The data also supports a model in which competitive DNA binding of different members of the NF-E2/AP1 transcription factor family modulates the developmental stage specificity of an erythroid enhancer. Feasibility to reswitch on embryonic/fetal globin genes through the manipulation of nuclear factor binding at a single regulatory DNA motif is discussed.
Keywords: transgenic mice/enhancer/passive repression/competitive factor binding/globin switch
Molecular and genetic data have indicated that the erythroid lineage-specific and developmental stage-specific expression of the human α- or β-globin gene family requires functional, and probably physical, interaction between the individual globin promoters with regulatory elements located far upstream of the individual gene clusters (reviewed in ref. 1). For the α-globin locus, the developmental regulation of the embryonic globin gene ζ (previously termed ζ2) and the two adult α-globin genes (α2 and α1) is controlled mainly by HS-40, an element located 40 kb upstream of ζ (2). Cis-linkage of HS-40 confers erythroid lineage-specific, autonomous, and appropriate developmental patterns of expression of either ζ- or α-globin promoter in transgenic mice (2–8), although the expression level of the human α-globin genes in adult mice is relatively low (4). Notably, HS-40 does not appear to confer position independence (8) and copy number dependence (4, 6–8) on the transgenic globin expression, with a sharp decrease of the promoter activity in transgenic mice when the copy numbers of the integrated transgenes are greater than two (4, 6, 7).
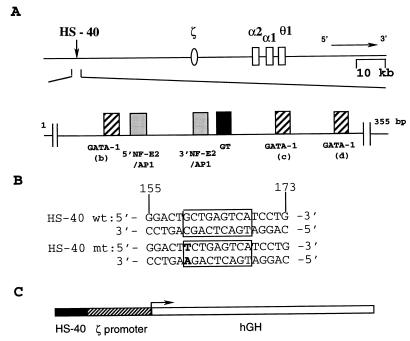
The regulatory signals for human globin switch appear to be transmitted through multiple protein–DNA complexes assembled at the globin promoters and the upstream regulatory elements (9). The functional domain of HS-40 has been mapped within a 300-bp stretch of DNA sequences, which consist of six nuclear factor-binding motifs (10) that are occupied in vivo in an erythroid lineage-specific and developmental stage-specific manner (11, 12). These include three GATA-1 motifs (b, c, and d), two NF-E2/AP1 motifs (5′ and 3′), and a GT motif (Fig. 1A). In vitro binding studies indicated that the GT motif predominately binds the ubiquitous Sp1 factor (13). The GATA-1 motifs bind the GATA family of transcription factors, including the erythroid-enriched GATA-1 (14). Finally, the NF-E2/AP1 motifs can bind several types of nuclear factors, including the erythroid-enriched NF-E2 (15) and the ubiquitous AP1 (16). Nrf1 (17–19), Nrf2 (20), and the small maf family of factors (ref. 21 and references therein) also participate in protein–DNA complex formation at this motif.
Figure 1.
(A) The linkage map of human α-like globin gene cluster. The nuclear factor-binding motifs are indicated below the map. (B) Nucleotide sequences of the wt and mt forms of the 3′NF-E2/AP1 motif of HS-40. (C) Map of the fragment used for microinjection of fertilized mouse eggs.
HS-40 first behaves as a classical enhancer for the ζ-globin as well as the α-globin promoters in transiently transfected erythroid cells (12, 22, 23). More systematic analysis by site-directed mutagenesis has revealed unexpected characteristics of HS-40 as a regulatory element (24, 25). Although most of the factor-binding motifs (Fig. 1A) positively regulate the HS-40 enhancer function, the 3′NF-E2/AP1 motif appears to exert both positive and negative regulatory effects on the ζ-globin promoter activity in embryonic/fetal K562 cells. Introduction of a 3-bp mutation into the central core of this motif prevents binding of both NF-E2 and AP1 and also lowers the HS-40 enhancer function by more than 75%. However, when a GC base pair at one boundary of the 3′NF-E2/AP1 binding site is changed to TA (3′NA-II, Fig. 1B), the HS-40 enhancer activity is increased by 2- to 3-fold (12). Because the same 1-bp mutation abolishes the binding of NF-E2, but not AP1, to this sequence (15, 26), we have hypothesized that transcriptional silencing of the human ζ-globin gene during transition from embryonic to fetal/adult erythroid development is, in conjunction with negative regulatory elements in the ζ-globin promoter (27), in part modulated by the competitive binding of positive regulatory factors such as AP1 and negatively regulatory NF-E2.
To test the physiological significance of this model, we have analyzed human ζ-globin promoter activity in transgenic mice, with the use of human growth hormone (hGH) as a reporter gene under the control of either wild-type (wt) HS-40 or its mutant (mt) form HS-40 (3′NA-II) (Fig. 1C). As shown below, our data strongly support the idea that silencing of the embryonic ζ-globin promoter during embryonic to fetal/adult erythroid development is passively regulated by HS-40 through the 3′NF-E2/AP1 motif. The study also has led to the discovery of a short, but powerful, erythroid-specific locus-control region (LCR) that could have interesting applications.
MATERIALS AND METHODS
Transgenic Mice.
Transgenic mice were produced by microinjection of DNA fragments into the pronuclei of fertilized mouse eggs (28, 29). Plasmids pHS40-ζ597-GH (ref. 12; abbreviated below as pHS40-ζGH) and pHS40 (3′NF-E2/AP1-II)-ζ597-GH [ref. 12; abbreviated below as pHS40 (3′NA-II)-ζGH], respectively, were cleaved with restriction enzymes EcoRI, NdeI, and ScaI. The 3.12-kb DNA fragments, HS40-ζGH and HS40(3′NA-II)-ζGH (Fig. 1C), were eluted from soft agarose gel, purified, and used for microinjection.
Transgenic founders were identified and their copy numbers of integrated transgenes were estimated by Southern blot analysis of the tail DNA. The founders then were bred with nontransgenic C57/B6 mice to establish lines. For analysis of expression in fetal (14.5 days postcoitum) and embryonic (9.5 days postcoitum) mice, transgenic males were mated to nontransgenic C57/B6 females. The morning on which the copulatory plug was observed was designated day 0.5. Transgenic pups were identified by PCR analysis of fetal mice tails or of embryo DNA. For each identification, duplicate PCRs were carried out by using one 5′ primer from the ζ-globin promoter region, and two different 3′ primers from the GH region.
GH Expression Assay.
The expression of the ζ-GH hybrid gene in adult mice was analyzed by GH assay (12, 30–33) and semiquantitative reverse transcription (RT)–PCR analysis (see below). For GH assay, blood samples were collected from the mouse tails. The levels of hGH in the blood then were quantitated with the Allego hGH radioimmunoassay kit from the Nichols Institute (San Juan Capistrano, CA) as described (12). When the concentrations of GH in the blood exceeded 50 ng/ml, the samples were diluted with horse serum to obtain a linear range for the GH assay.
RT-PCR.
To analyze the RNA levels at fetal and embryonic stages, liquid N2-frozen 9.5-day embryos, 14.5-day fetuses, or 14.5-day fetal livers were manually homogenized, and the RNAs were isolated by acid guandinium isothiocyanate/phenol/chloroform extraction (34). For adult samples, the mice were rendered anemic by three injections of phenylhydrazine (40 μg/g of body weight) at 0, 8, and 24 hr. Six days after the first infection, the mice were sacrificed, and RNAs were isolated from different tissues as described above. In all cases, the total RNAs were used for assay without further purification.
RT-PCRs were carried out as described (35, 36). Each RT reaction mixture contained 1 μg of RNA, 200 units of Superscript II reverse transcriptase (GIBCO/BRL), and 20 mM oligo(dT)15 as the primer. One-twentieth of cDNAs then were PCR-amplified with Taq polymerase (GIBCO/BRL) and primers specific for hGH, mouse mβmajor, mζ, or mouse glyceraldehyde-3-phosphate dehydrogenase (mG3PDH). All amplifications were carried out in a Hybrid OmniGene system with the following temperature profiles: an initial denaturation at 95°C for 3 min, 53°C for 1 min, and 72°C for 1 min; followed by repeating renaturation cycles of 95°C for 1 min, 53°C for 1 min, and 72°C for 1 min, and finally an elongation step at 72°C for 5 min. Each PCR analysis was done in duplicate. The sequences of PCR primers used were: 5′mG3PDH, 5′-TGAAGGTCGGTGTGAACGGATTTGGC-3′; 3′mG3PDH, 5′-CATGTAGGCCATGAGGTCCACCAC-3′; 5′hGH, 5′-GTCCCTGCTCCTGGCTTT-3; 3′hGH, 5′-ATGCGGAGCAGCTCCAGGTT-3′ and 5′-CATCAGCGTTTGGATGCCTT-3′; 5′-mβmajor, 5′-TGGGCAGGCTGCTGGTTA-3′; 3′mβmajor, 5′-TTAGTGGTACTTGTGAGCCAA-3′; 5′mζ, 5′-CTGATGAAGAATGAGAGAGC-3′; 3′mζ, 5′-TAGAGGTACTTCTCATCAGTCAG-3′. The PCR band lengths were 980 bp for mG3PDH, 335 bp for mβmajor, and 290 bp or 450 bp for ζ-GH. One-fifth of each PCR product was resolved on a 1.5% agrose-ethidium bromide gel, which was then documented by the IS1000 Digital Imaging System and saved in computer tif format. The band intensities then were quantitiated with the PhosphorImage system.
For semiquantitative purposes, mG3PDH was used as the internal standard. The linearity of amplification of the G3PDH cDNA first was defined by PCR of series dilutions of the cDNAs. The cycle number of 25 was chosen for amplifying mG3PDH, because under the reaction conditions described above it resulted in signal linearity over a range of the serial dilutions of different mouse tissue cDNAs. In the initial calibration test, mG3PDH bands with similar intensities were obtained from the different tissue cDNAs when the same amounts of RNAs were used for RT. The appropriate PCR cycle numbers used to amplify the hGH, mβmaj, and mζ transcripts were 28, 25, and 28, respectively. Coamplification of different cDNAs from different genes was tested to be unsuccessful, presumably because of random primer competitions. Thus, the amounts of different tissue cDNAs used first were determined by PCR using the mG3PDG primers, then individual PCRs using the hGH, mβmaj, or mζ primers were carried out.
RESULTS
Position Independence and Copy Number Dependence of Human ζ-Globin Promoter Activities in Adult HS40(3′NA-II)-ζGH Mice.
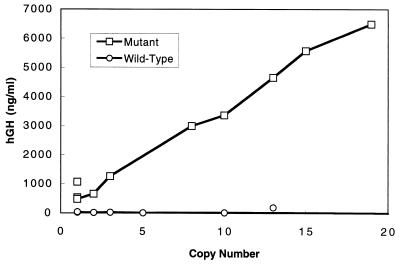
A total of nine founders with the wt HS40-ζGH transgene and 10 founders with the mt HS40(3′NA-II)-ζGH transgene have been obtained. The copy numbers of integrated fragments in these mice vary from one to more than 100 (Table 1). The ζ-globin promoter activities in the founder mice first were measured with the sensitive hGH assay. The hGH previously has been used successfully as a reporter gene in DNA transfection and transgenic mice experiments. The amounts of secreted enzyme molecules are good representations of the quantities of mRNAs inside the expressing cells (refs. 12 and 30–33 and references therein). As shown in Table 1 and Fig. 2, except for founder 100A, all of the blood GH amounts per copy of integrated transgenes were very low or comparable to the nontransgenic controls (data not shown). This result is consistent with previous findings by others that human ζ-globin promoter activity is essentially shut off in adult transgenic mice, even when it is linked in cis with the HS-40 element (5, 8) or with β-LCR (37, 38).
Table 1.
Copy numbers and adult expression levels of transgenic mice
| Mutant type
|
Wild type
|
||||
|---|---|---|---|---|---|
| Founder line | Copy number | hGH, ng/ml | Founder line | Copy number | hGH, ng/ml |
| 1A* | 1 | 470 | 1A* | 1 | 36 |
| 1B* | 1 | 530 | 1B* | 1 | 20 |
| 1C* | 1 | 1,060 | 2 | 2 | 14 |
| 2 | 2 | 650 | 3 | 3 | 22 |
| 3 | 3 | 1,260 | 5 | 5 | 5 |
| 8* | 8 | 2,990 | 10* | 10 | 13 |
| 10* | 10 | 3,360 | 13* | 13 | 187 |
| 13* | 13 | 4,650 | 100A | >100 | 1,400 |
| 15* | 15 | 5,560 | 100B | >100 | 30 |
| 19* | 19 | 6,490 | |||
The designations of founder mice integrated with wt HS40-ζGH or mt HS40(3′NA-II)-ζGH are listed in the first columns. Those founders for which lines have been established are indicated by ∗. The copy numbers of the transgenes are listed in the second columns. The levels of GHs in the sera of adult mice treated with phenylhydrazine are listed in the third columns. For hGH analysis, the mice with wt transgene were all assayed at the age of 5 months except founder 1B, which was 9 months old. Mice with the mt transgene were all assayed at the age of 4 months except founder 15, which was 2 months old.
Figure 2.
Copy number-dependence of transgene expression of the ζ-GH hybrid gene cis-linked with the wt or the mt HS-40. The expression levels were determined by hGH assay of blood samples as listed in Table 1.
On the contrary, the blood hGH amounts of the 10 mt founders, when measured at the ages of 2–4 months, exhibited a nearly straight line as a function of the copy numbers of integrated genes. The data indicate that when under the control of the mt HS40 element the ζ-GH gene is actively transcribed in the adult mice, and the expression is in general copy number dependent and integration site independent. The blood GH assay of these founders at other ages as well as of their progenies supports the above general conclusion, although with somewhat variable quantities of hGH depending on the ages of the mice and the growth conditions (data not shown).
Adult Erythroid Tissue Specificities of ζ-GH Gene Expression.
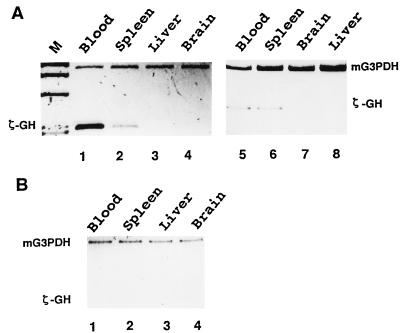
In the developing mouse, the first site of erythropoiesis is at the yolk sac blood island during 8–14 days of gestation. The major site of erythropoiesis then shifts to the fetal liver, and finally to the spleen at birth. We have examined by RT-PCR the expression patterns of the human ζ-GH hybrid gene in different tissues of adult transgenic mice. In all cases analyzed, the transcription of ζ-GH in the mt mice lines appears to be restricted to the erythroid tissues. As shown in Fig. 3A for mt 15 and 1C, the ζ-GH transgene is expressed mainly in the spleens and blood of anemic mice (Fig. 3A, lanes 1, 2, 5, and 6), but not in their livers and brains (Fig. 3A, lanes 3, 4, 7, and 8). Analysis of adult mice from mt lines 8 and 19 gives similar results, and RT-PCR signals of ζ-GH could not be observed without phenylhydrazine treatment of the mt mice (data not shown). Also consistent with the GH assay is the small amount or absence of the human ζ-GH transcripts in tissues of wt transgenic mice, as exemplified for wt line 1B (Fig. 3B).
Figure 3.
Expression of ζ-GH hybrid gene in adult transgenic mice. The levels of ζ-GH transcripts in different tissues of transgenic mice were estimated by RT-PCR. The signals from mG3PDH were used as the references. (A) Left, mt line 15; Right, mt line 1C. M, HaeIII digested φ ×174 DNA as the length marker. (B) wt line 1B.
Transgenic Expression During Development.
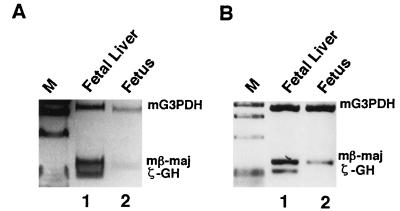
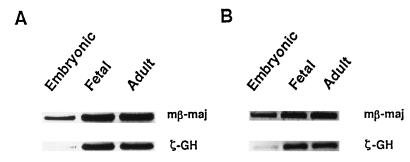
The expression of transgenic ζ-GH at the fetal stage also appears to be erythroid specific. As shown in Fig. 4, ζ-GH transcripts could be detected in 14.5-day fetuses from transgenic mice with either mt HS40-ζGH (Fig. 4A, line 15) or wt HS40-ζGH (Fig. 4B, line 13), but not in the nontransgenic controls (data not shown). Furthermore, the higher intensity of the RT-PCR band derived from the fetal liver RNA (Fig. 4, lanes 1) than that from the whole fetus (Fig. 4, lanes 2) is consistent with the erythroid fetal liver being the major site of transcription of the ζ-GH transgenes of the fetuses. Similar conclusions were reached from analysis of mt lines 10 and 19 and wt line 1A (data not shown).
Figure 4.
Expression of ζ-GH hybrid gene of fetal transgenic mice. RNAs isolated from the fetal livers (lanes 1) or whole fetuses (lanes 2) were analyzed by RT-PCR. (A) mt line 15. (B) wt line 13. Note that to visualize the ζ-GH signal better 3-fold more sample of the RT-PCR product from ζ-GH transcripts was loaded in lane 1 of B.
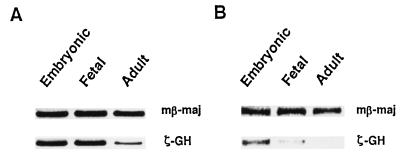
The changes of ζ-GH RNA levels during mouse development also have been followed by RT-PCR, and examples of the analysis are shown in Fig. 5. In general, wt HS40 confers the correct temporal patterns of expression of ζ-GH during development, with the level of ζ-GH transcripts relatively high in 9.5-day embryo, but drops significantly in the adult blood (Fig. 5B). On the other hand, consistent with the GH assay data of Fig. 2, the activity of the human ζ-globin promoter cis-linked with the mt HS-40 (3′NA-II) is indeed derepressed. As shown in Fig. 5A for single-copy mt 1C, the ζ-GH transcript levels at all three stages are significantly more abundant than the single-copy wt 1A (Fig. 5B). Interestingly, the ζ-GH expression of multiple-copy mt 15 and mt 19, while derepressed at the adult stage, is greatly repressed in the embryos (Fig. 6).
Figure 5.
Expression of ζ-GH hybrid gene during development. RNAs isolated from embryonic yolk sac, fetal liver, and adult blood were analyzed by RT-PCR for mt line 1C (A) and wt line 1A (B).
Figure 6.
RT-PCR analysis of ζ-GH hybrid gene expression in multicopy/transgenic lines mt 15 (A) and mt 19 (B).
DISCUSSION
The element HS-40 is known as a developmental stage-specific ehnancer for the expression of human ζ- and α-globin promoter activities in embryonic and adult erythroid cells, respectively. In this transgenic mice study, we have demonstrated that a single bp mutation of HS-40 converts the element into a potent LCR for derepression of the ζ-globin promoter in adult and fetal erythroid cells.
Transcription of the ζ-GH transgenes is erythroid tissue specific (Figs. 3 and 4). However, the ζ-GH transcripts are not accumulated and retained in the reticulocytes (data not shown). This situation is similar to other reporter gene mRNAs, and it is most likely caused by their instability in the reticulocytes (S. Liebhaber, personal communication). However, based on previous transgenic mice studies of GH under the control of metallothione promoter (30, 31), we could make a rough estimation of the population size of ζ-GH mRNA in the adult transgenic mice. Assuming similar efficiencies of translation of the metallothione in GH and ζ-GH hybrid transcripts and a range of 105–107 red cells at the proerythroblast through orthochromatic stage (refs. 39 and 40 and references therein), it would appear that as many as several hundred to several thousand ζ-GH mRNAs exist in each expressing adult erythroid cell (calculation not shown).
Although our RNA analysis data of the transgenic mice are only semiquantitative and few embryo samples have been analyzed in comparison to the adult blood samples, together with the GH assay they provide several interesting and intriguing implications. First, regarding the molecular mechanisms of human ζ- to α-globin switch during erythroid development, it has been documented that the HS-40 element is both necessary and sufficient for the appropriate developmental regulation of ζ-globin promoter activity in erythroid cells, on intact human chromosome 16 and in transgenic mice (refs. 5, 6, 8, and 41 and references therein). Furthermore, similar to the autonomous regulation of the human embryonic ɛ-globin gene by the β-LCR (42, 43), failure of successful competition against the α-globin promoters for a functional interaction with HS-40 does not seem to be responsible for the on-and-off transition of ζ-globin expression during development (5, 8). On the other hand, in transient DNA transfection experiments, two nuclear factor-binding motifs appear to be involved in the negative regulation of ζ-globin promoter during embryonic-to-fetal transition of the erythroid cells (27). Consistent with these studies, in at least eight of nine mouse founders/lines carrying the human ζ-GH hybrid gene cis-linked with the wt HS-40, the human ζ-globin promoter is relatively inactive at the adult stage, irrespective of the copy numbers of the transgenes (Table 1, Figs. 2 and 3). However, introduction of the single bp mutation into the 3′NF-E2/AP1 motif of HS-40 significantly derepresses human ζ-globin promoter activity in the adult and fetal erythroid lineages (Table 1, Figs. 3–5). As estimated above from the GH assay data, in adult mice the expression level of the ζ-GH gene under control of the mt HS-40 is comparable to those directed by potent promoters such as that of the metallothione in gene. Some derepression also may occur in embryos of transgenic lines carrying a single copy of the mt construct (Fig. 5).
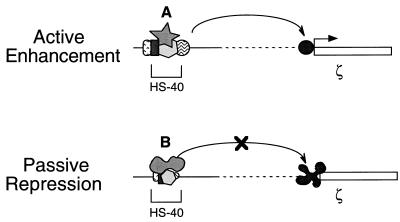
Combining the transgenic mouse data from this study, the previous DNA transfection experiments in K562 cells (24, 25), and the consequences of the 3′NA-II mutation on nuclear factor binding (refs. 15 and 26 and references therein), we propose a model in which competitive binding of nuclear factors at a single DNA motif within an enhancer modulates its dual functional roles during development: active enhancement and passive repression (Fig. 7). It appears that the adult and fetal erythroid cells contain a set of nuclear factor components, DNA binding and non-DNA binding, necessary for the activation of the ζ-globin promoter. However, among several candidate factors (16–21), the dominant binding by a factor such as NF-E2 at the 3′NF-E2/AP1 motif of HS-40 allows the formation of a specific multiple protein–DNA complex. This complex could functionally interact and activate the α-globin promoters in adult erythroid cells, but it could not overcome the negative regulatory mechanisms silencing the ζ-globin promoter (Fig. 7, passive repression). However, when the NF-E2 binding at this motif of the HS-40 is inhibited, or conformationally modified such as by mutation of the 3′NF-E2/AP1 motif or through change of the effective concentration of NF-E2 relative to a modified form of NF-E2 or another factor such as AP1, binding of the latter factor(s) takes over and a different complex of HS-40 is formed. This new HS-40 complex would reactivate the silenced ζ-globin promoter in the adult erythroid cells (Fig. 7, active enhancement). Presumably, the conformation of this latter complex is functionally similar to that in the embryo yolk sac, which preferentially activates the ζ-, but not α-, globin promoter. The model provides an interesting scenario in which the physical architecture and the associated function of an enhancer could be developmentally modulated through the manipulation of nuclear factor binding at a single DNA motif. Whether a similar regulatory scenario occurs at the NF-E2/AP1 motifs of the β-LCR, and the potential therapeutic use of this finding in manipulating the human globin switch, await to be investigated.
Figure 7.
A cartoon model of the HS-40 function. It is proposed that the multiple protein–DNA complex(es) formed within HS-40 adopts different conformations in the embryonic and adult erythroid cells. This conformational difference results at least in part from competitive binding between different nuclear factors, A and B, at the 3′NF-E2/AP1 motif. In the upper scheme, the HS-40 enhancer would activate transcription of the ζ-globin gene through interaction with its promoter. On the other hand, the altered HS-40 complex shown in the lower scheme is unable to overcome the negative regulatory effect of repressing elements in the ζ-globin promoter. See text for more discussion.
Interestingly, the mt HS-40 (3′NA-II) behaves as a potent, erythroid-specific LCR for expression of the human ζ-globin promoter in adult erythroid cells. The levels of expression of the integrated HS-40 (3′NA-II)-ζ GH transgenes continue to increase even at the copy numbers of 19. In particular, founder mice of similar ages exhibited a linear dependence on the blood GH levels as a function of the transgene copies (Table 1 and Fig. 2). This finding is in great contrast to previous transgenic mice studies of the transcriptional activation by wt HS-40 of the human ζ- or α-globin promoter (4–8). The expression per copy of the tandemly arranged transgenes sharply decreases even when the copy number exceeds two per genome (4, 6). This phenomenon is common in many other transgenic mice experiments as well, and the repression has been interpreted as the result of a cis-acting effect from the heterochomatin structure associated with the tandem array of the transgenes (4, 44). The human β-globin-LCR, while being one of the most potent LCRs known (refs. 1 and 46 and references therein), appears to function up to the copy number of 5–9 integrated transgenes in mice (45). The HS-40 (3′NA-II)-ζ promoter thus may provide a convenient enhancer/promoter system for high-level expression of foreign genes in the adult/fetal erythroid cells of transgenic animals.
Accompanied with the above heterochromatin effect caused by tandem transgene arrangement, the activities of many promoters, including the human ζ-globin promoter linked in cis with the wt HS-40, in transgenic mice are highly variable among different lines (8, 46). As deduced from studies of the position effect variegation in yeast and Drosophila (reviewed in refs. 47 and 48), this position-dependent variation of expression in mammalian cells most likely results from influence of the chromatin environment near the sites of transgene integration (discussed in ref. 49 and references therein). The human α-globin locus adopts an open chromatin configuration in both erythroid and nonerythroid cells, whether or not the HS-40 complex is formed. It is thus not surprising that the wt HS-40 element lacks a function to overcome the repressing effect from the environmental chromatin structure. Although the molecular basis for the mt HS-40(3′NA-II)’s ability to confer position independence on human ζ-globin promoter activity is unclear, this property in conjunction with its relatively small size makes the HS-40(3′NA-II)-ζ promoter an attractive element for use for viral vector-based gene expression experiments in erythroid and possibly certain hematopoietic progenitor cells.
Acknowledgments
We thank our colleagues for help and discussions. We also thank Doug Engle for his helpful suggestions on the semiquantitative PCR protocols. This research was supported by Academia Sinica (Republic of China, ROC), National Health Research Institute (ROC), National Science Council (ROC), Foundation of Biomedical Sciences (ROC), and National Institutes of Health Grant DK 29800.
ABBREVIATIONS
- GH
growth hormone
- hGH
human growth hormone
- G3PDH
glyceraldehyde-3-phosphate dehydrogenase
- LCR
locus-control region
- RT
reverse transcription
- wt
wild type
- mut
mutant
References
- 1.Grosveld F, Dillon N, Higgs D. Baillieres Clin Haematol. 1993;6:31–55. doi: 10.1016/s0950-3536(05)80065-4. [DOI] [PubMed] [Google Scholar]
- 2.Higgs D R, Wood W G, Jarman A P, Sharpe J, Lida J, Pretorius I-M, Ayyub H. Genes Dev. 1990;4:1588–1601. doi: 10.1101/gad.4.9.1588. [DOI] [PubMed] [Google Scholar]
- 3.Vyas P, Vickers M A, Simmons D L, Ayyub H, Graddock C F, Higgs D R. Cell. 1992;69:781–793. doi: 10.1016/0092-8674(92)90290-s. [DOI] [PubMed] [Google Scholar]
- 4.Sharpe J A, Chen-Thomas P S, Lida J, Ayyub H, Wood W G, Higgs D R. EMBO J. 1992;11:4565–4572. doi: 10.1002/j.1460-2075.1992.tb05558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pondel M D, Proudfoot N J, Whitelaw C, Whitelaw E. Nucleic Acids Res. 1992;20:5655–5660. doi: 10.1093/nar/20.21.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharpe J A, Wells D J, Whitelaw E, Vyas P, Higgs D R, Wood W G. Proc Natl Acad Sci USA. 1993;90:11262–11266. doi: 10.1073/pnas.90.23.11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gourdon G, Sharpe J A, Wells D, Wood W G, Higgs D R. Nucleic Acids Res. 1994;22:4139–4147. doi: 10.1093/nar/22.20.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robertson G, Garrick D, Wu W, Kearns M, Martin D, Whitelaw E. Proc Natl Acad Sci USA. 1995;92:5371–5375. doi: 10.1073/pnas.92.12.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stamatoyannopoulos G, editor. Molecular Biology of Hemoglobin Switching. Andover, Hampshire, U.K.: Intercept Limited; 1995. [Google Scholar]
- 10.Jarman A P, Wood W G, Sharp J A, Gourdon G, Ayyub H, Higgs D R. Mol Cell Biol. 1991;11:4679–4689. doi: 10.1128/mcb.11.9.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strauss E C, Andrews N C, Higgs D R, Orkin S H. Mol Cell Biol. 1992;12:2135–2142. doi: 10.1128/mcb.12.5.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q, Reddy P M S, Yu C Y, Bastiani C, Higgs D, Stamatoyannopoulos G, Papayannopoulou T, Shen C-K J. Mol Cell Biol. 1993;13:2298–2308. doi: 10.1128/mcb.13.4.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Courey A J, Tjian R. Cell. 1998;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 14.Martin D I K, Zon L I, Mutter G, Orkin S H. Nature (London) 1990;344:444–447. doi: 10.1038/344444a0. [DOI] [PubMed] [Google Scholar]
- 15.Andrews N C, Erdjument-Bromage H, Davison M B, Tempst P, Orkin S H. Nature (London) 1993;362:722–728. doi: 10.1038/362722a0. [DOI] [PubMed] [Google Scholar]
- 16.Lee W, Haslinger A, Karin M, Tjian R. Nature (London) 1987;325:368–372. doi: 10.1038/325368a0. [DOI] [PubMed] [Google Scholar]
- 17.Chan J Y, Han X-L, Kan Y W. Proc Natl Acad Sci USA. 1993;90:11371–11375. doi: 10.1073/pnas.90.23.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caterina J J, Donze D, Sun C-W, Ciavatta D J, Townes T M. Nucleic Acids Res. 1994;22:2383–2391. doi: 10.1093/nar/22.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luna L, Johnsen O, Skartlien A H, Pedeutour F, Turc-Carel C, Prydz H, Kolsto A-B. Genomics. 1994;22:553–562. doi: 10.1006/geno.1994.1428. [DOI] [PubMed] [Google Scholar]
- 20.Moi P, Chan K, Asunis I, Cao A, Kan Y W. Proc Natl Acad Sci USA. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kataoka K, Igarashi K, Itoh K, Fujiwara K T, Noda M, Yamamoto M, Nishizawa M. Mol Cell Biol. 1995;15:2180–2190. doi: 10.1128/mcb.15.4.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pondel M D, George M, Proudfoot N J. Nucleic Acids Res. 1992;20:237–243. doi: 10.1093/nar/20.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren S, Luo X-N, Atweh G F. Blood. 1993;81:1058–1066. [PubMed] [Google Scholar]
- 24.Zhang Q, Rombel I, Reddy G N, Gang J-B, Shen C-K J. J Biol Chem. 1995;270:8501–8505. doi: 10.1074/jbc.270.15.8501. [DOI] [PubMed] [Google Scholar]
- 25.Rombel I, Hu K-Y, Zhang Q, Papayannopoulou T, Stamatoyannopoulos G, Shen C-K J. Proc Natl Acad Sci USA. 1995;92:6454–6458. doi: 10.1073/pnas.92.14.6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ney P A, Sorrentino B P, McDonagh K T, Nienhuis A W. Genes Dev. 1990;4:993–1006. doi: 10.1101/gad.4.6.993. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Q, Rombel I, Reddy G N, Shen C-K J. In: Molecular Biology of Hemoglobin Switch. Stamatoyannopoulos G, editor. Andover, Hampshire, U.K.: Intercept Limited; 1995. pp. 193–202. [Google Scholar]
- 28.Brinster R L, Chen H, Trumbauer M, Senear A W, Warren R, Palmiter R D. Cell. 1981;27:223–231. doi: 10.1016/0092-8674(81)90376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costantini F, Lacy E. Nature (London) 1981;294:92–94. doi: 10.1038/294092a0. [DOI] [PubMed] [Google Scholar]
- 30.Palmiter R D, Brinster R L, Hammer R E, Trumbauer M E, Rosenfeld M G, Brinberg N C, Evans R M. Nature (London) 1982;300:611–615. doi: 10.1038/300611a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palmiter R D, Norstedt G, Gelinas R E, Hammer R E, Brinster R L. Science. 1983;222:809–814. doi: 10.1126/science.6356363. [DOI] [PubMed] [Google Scholar]
- 32.Hammer R E, Pursel V G, Rexroad C E, Wall R J, Bolt D J, Ebert K M, Palmiter R D, Brinster R L. Nature (London) 1985;315:680–683. doi: 10.1038/315680a0. [DOI] [PubMed] [Google Scholar]
- 33.Selden R F, Howie K B, Rowe M E, Goodman H M, Moore D D. Mol Cell Biol. 1986;6:3173–3179. doi: 10.1128/mcb.6.9.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 35.Chelly J, Kaplan J-C, Maire P, Gautron S, Kahn A. Nature (London) 1988;333:858–860. doi: 10.1038/333858a0. [DOI] [PubMed] [Google Scholar]
- 36.Foley K P, Leonard M W, Engel J D. Trends Genet. 1993;9:380–385. doi: 10.1016/0168-9525(93)90137-7. [DOI] [PubMed] [Google Scholar]
- 37.Albitar M, Katsumata M, Liebhaber S A. Mol Cell Biol. 1991;11:3786–3794. doi: 10.1128/mcb.11.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spangler E A, Andrews K A, Rubin E M. Nucleic Acids Res. 1990;18:7093–7097. doi: 10.1093/nar/18.23.7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Broudy V C, Lin N L, Priestley G V, Nocka K, Wolf N S. Blood. 1996;88:75–81. [PubMed] [Google Scholar]
- 40.Cheng T-C, Polmar S K, Kazazian H H. J Biol Chem. 1974;249:1781–1786. [PubMed] [Google Scholar]
- 41.Nicholls R D, Fischel-Ghodsian N, Higgs D R. Cell. 1987;49:369–378. doi: 10.1016/0092-8674(87)90289-3. [DOI] [PubMed] [Google Scholar]
- 42.Shih D M, Wall R J, Shapiro S G. Nucleic Acids Res. 1990;18:5465–5472. doi: 10.1093/nar/18.18.5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raich N, Enver T, Nakamoto B, Josephon B, Papayannopoulou T, Stamatoyannopoulos G. Science. 1990;250:1147–1149. doi: 10.1126/science.2251502. [DOI] [PubMed] [Google Scholar]
- 44.Sabl J F, Henikoff S. Genetics. 1996;142:447–458. doi: 10.1093/genetics/142.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ellis J, Tan-Un K C, Harper A, Michalovich D, Yannoutsos N, Philipsen S, Grosveld F. EMBO J. 1996;15:562–568. [PMC free article] [PubMed] [Google Scholar]
- 46.Guy L-G, Kothary R, Wall L. Nucleic Acids Res. 1997;25:4400–4407. doi: 10.1093/nar/25.21.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henikoff S. Curr Opin Genet Dev. 1992;2:907–912. doi: 10.1016/s0959-437x(05)80114-5. [DOI] [PubMed] [Google Scholar]
- 48.Pirrotta V, Rastelli L. BioEssays. 1994;16:549–556. doi: 10.1002/bies.950160808. [DOI] [PubMed] [Google Scholar]
- 49.Walters M C, Magis W, Fiering S, Eidemiller J, Scalzo D, Groudine M, Martin D I K. Genes Dev. 1996;10:185–195. doi: 10.1101/gad.10.2.185. [DOI] [PubMed] [Google Scholar]