Abstract
To determine the dynamics of transcript extrusion from Escherichia coli RNA polymerase (RNAP), we used degradation of the RNA by RNases T1 and A in a series of consecutive elongation complexes (ECs). In intact ECs, even extremely high doses of the RNases were unable to cut the RNA closer than 14–16 nt from the 3′ end. Our results prove that all of the cuts detected within the 14-nt zone are derived from the EC that is denatured during inactivation of the RNases. The protected zone monotonously translocates along the RNA after addition of new nucleotides to the transcript. The upstream region of the RNA heading toward the 5′ end is cleaved and dissociated from the EC, with no effect on the stability and activity of the EC. Most of the current data suggest that an 8- to 10-nt RNA⋅DNA hybrid is formed in the EC. Here, we show that an 8- to 10-nt RNA obtained by truncating the RNase-generated products further with either GreB or pyrophosphate is sufficient for the high stability and activity of the EC. This result suggests that the transcript–RNAP interaction that is required for holding the EC together can be limited to the RNA region involved in the 8- to 10-nt RNA⋅DNA hybrid.
The regulation of transcription elongation is a significant element of genome expression in both prokaryotes and eukaryotes (1). To explain the dramatic sequence-dependent changes in the rate of transcription during pausing and arrest, and in the stability of the elongation complex (EC) during termination (2–4), a detailed understanding of the structure of the elongation intermediates is required.
Many of the signals regulating transcription are intrinsically encoded in the nascent RNA (5). There are three important aspects of transcript arrangement in RNA polymerase (RNAP). The first is the point at which the RNA emerges from the enzyme. Once freed from RNAP, the RNA is capable of forming stem-loop structures that participate in termination, antitermination, and pausing of transcription (3, 5, 6). At this point, the RNA also becomes available for binding accessory factors. Proteins such as Rho, NusA, and NusB interact with nascent RNA to regulate pausing and termination (3–6). Studies of mixed populations of ECs have shown that 12 ± 2 nt in the transcript are protected from single-strand-specific RNases, suggesting that this part of the RNA is covered by RNAP or is involved in a hybrid with the template (7). In the individual ECs of Escherichia coli RNAP, cleavages with low doses of RNases A and T1 were substantially inhibited within ≈16 nt of the 3′-proximal RNA (8). In the active ECs of mammalian RNAP II, the zone protected from RNase P1 comprises 17–20 nt (9). The implication of a self-cleavable hammerhead RNA that is newly synthesized by RNAP and that subsequently cleaves itself out of the transcript in the EC has revealed that folding of the RNA into the secondary structure is allowed as close as 9–12 nt from the 3′ end of the transcript (10). On the other hand, crosslinking experiments have proved that the RNA remains in contact with the protein at much longer distances. A crosslink between RNAP and a ribonucleotide analog incorporated into the RNA was formed when the analog was moved to distances up to 90 nt from the growing end of the RNA (11).
The second aspect of RNA arrangement in RNAP is the extent and role of complementary interactions between the nascent RNA and the DNA. In the classic view, the 3′-proximal part of the RNA and the template DNA strand form a 12-bp hybrid in the transcription bubble (2, 12). This idea was based on thermodynamic analysis of the elongation process and on RNA footprinting data (7, 13). The experiments with crosslinkable ribonucleotide analogs also demonstrated a close proximity between 7 and 10 nt at the 3′ end of the RNA and the template, which was considered by the authors as proof of the existence of the hybrid (14). A strong argument opposing the crosslinking data was provided by the observation of the cleavage of the RNA in some ECs as close as 3 nt from the growing tip of the RNA with high doses of single-strand-specific RNases. This finding contradicted the idea of extended RNA–DNA pairing and gave rise to the alternative model that accepts the 2- to 3-bp hybrid (15, 16).
The third aspect of RNA arrangement in RNAP is the specificity of the transcript–protein contacts outside the RNA⋅DNA hybrid and the contribution of these interactions to the catalytic activity and stability of the EC. Initially, the strength of the 12-bp hybrid was thought to determine the stability of the EC (2, 13). In this view, the weakness of the oligo(U)⋅oligo(dA) hybrid was considered a major cause of the dissociation of the EC at transcription terminators (2). However, the importance of protein–RNA interactions also has been noted in a number of studies showing that RNAP forms catalytically active binary complexes with RNA in the absence of template (17). The idea of a special RNA-binding site in RNAP that is responsible for EC stability is supported by the RNase data in favor of a 3-nt hybrid, because such a short hybrid is insufficiently stable to keep the RNA associated with the ternary complex (2, 16). In addition, the stable complexes containing very short 4- to 6-nt RNAs were obtained on E. coli PL and rrnB P1 promoters during reiterative RNA synthesis, which further argued that additional stabilization power should come to the EC from the interaction of RNA with the protein (18, 19).
The goals of our current study were to analyze the contacts between E. coli RNAP and the nascent RNA and to determine the minimal length of RNA that is sufficient for the high stability of the EC. This analysis complements a number of previous studies that were concentrated mostly on DNA–protein relationships in ECs at different positions along the template (16, 20, 21).
MATERIALS AND METHODS
Transcription Template and Transcription Reactions.
His-tagged RNAP was purified and immobilized on Ni2+-nitrilotriacetate (NTA)-agarose (Qiagen) as described (20). RNA-labeled immobilized ECs that were halted at different positions of the DNA template containing T7A1 promoter were obtained as described (21) except that ApUpC (100 μM) was used as the primer. The homogeneous arrested complex was purified by adding all four NTPs (2.5 mM each for a 10-min chase) followed by washing with transcription buffer (TB: 20 mM Tris⋅HCl, pH 7.9/40 mM KCl/5 mM MgCl2/1 mM 2-mercaptoethanol) containing 1 M KCl (21). All procedures were performed at 24°C, unless otherwise indicated.
Cleavage of the RNA by RNases A and T1.
Several samples of the immobilized EC in 10 μl of TB were incubated for 10 min with different concentrations of RNase A (Sigma) or RNase T1 (Boehringer Mannheim) as specified in the figure legends. After RNase treatment, one of the samples was combined with 3 μl of phenol (the “total” fraction) or with 20 μl of SDS-loading buffer (7 M urea/10 mM EDTA/0.5% SDS) and vortexed immediately for 5 sec. To test the integrity of the EC after cleavage, another sample was combined with 10 μl of TB, and, after a brief centrifugation, 10 μl of the supernatant was removed and 3 μl of phenol was added as described above (the “supernatant” fraction). Ten microliters of the remaining pellet was also combined with 3 μl of phenol (the “pellet” fraction). To obtain the “pellet-washed” fraction, the cleaved complex was washed three times with 1 ml of TB, and 3 μl of phenol was added. To test the catalytic activity of the EC, the cleaved complex was incubated with 50 μM NTP for 30 sec and the RNase was inactivated by adding 3 μl of phenol (the “chase”). Where indicated, guanosine 2′-monophosphate (2′-GMP; Sigma) at a final concentration of 100 mM, or 6 μl of Prime RNase Inhibitor (5 Prime → 3 Prime) were added before the addition of phenol. All samples were combined with 10 μl of gel loading buffer (50 mM EDTA/10 M urea) and separated on denaturing urea/PAGE.
Test for EC Stability.
Twenty microliters of washed immobilized EC, containing either full-sized or truncated RNA, was incubated for 30 min in TB containing 300 mM KCl. After a brief centrifugation, 10 μl of the supernatant was removed and combined with an equal volume of gel loading buffer. The remaining pellet also was combined with an equal volume of gel loading buffer.
GreB-Induced RNA Cleavage and Pyrophosphorolysis.
The ECs were incubated for 30 min with 5,000 units/ml RNase T1 and washed 10 times with 1 ml of TB. GreB protein was purified as described (22). The truncated ECs were treated with 0.5 mg/ml GreB or with 2.5 mM sodium pyrophosphate (PPi; Fisher) for 30 min. Excess GreB or PPi was removed by washing the EC samples five times with 1 ml of TB. The ECs then were tested for their stability and for their ability to elongate the RNA (50 μM NTP each for a 30-sec chase) as described above.
RESULTS AND DISCUSSION
Development of a Method of RNA Truncation in Intact ECs by RNases T1 and A.
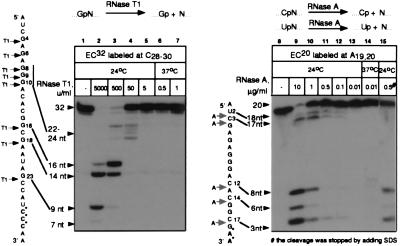
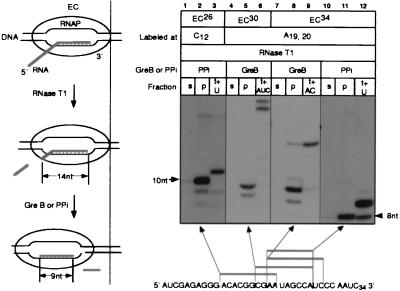
To study the contacts between RNAP and the transcript, we used RNA-labeled ECs immobilized in a solid phase. The complexes were obtained from the A1 promoter of T7 phage and “walked” to the destination site by alternating the subsets of NTPs added to the reaction (20, 23, 24). We treated the ECs with RNase T1 or A, which cleave the phosphate linkage after guanine and pyrimidine residues, respectively (see the scheme in Fig. 1). Both RNases are specific for single-stranded RNA and generate products bearing the phosphate at the 3′ end. EC32 and EC20 were selected to elaborate the conditions for treatment with RNases T1 and A, respectively (the numerical index denotes the length of the RNA in nontreated EC).
Figure 1.
Treatment of ECs with increasing doses of RNase T1 or A. RNA-labeled EC32 or EC20 was incubated with the indicated concentrations of RNase T1 or A, respectively, at the temperatures indicated. Cleavage was stopped with the addition of phenol (lanes 2–7 and 9–14) or SDS (lane 15). Here and in the other figures, RNA sequences are shown next to the autoradiographs; asterisks mark the positions of labeling; arrows show the cuts introduced by the RNases; and numerical indexes indicate the positions of the residues starting from the 5′ end of the transcript. Numbers and arrows show the length of the RNA.
When the concentration of RNase T1 was increased, the RNA in EC32 was truncated from the 5′ end—first to 22–24 nt, and then to 14–16 nt (Fig. 1, lanes 3–5). The highest dose of RNase T1 (5,000 units/ml) caused cleavage at the G23 residue (as counted from the start point of transcription), located 9 nt from the 3′ end of the RNA (lane 2). Treatment of EC20 with a stepwise increase in the amount of RNase A caused a gradual reduction of the size of labeled RNA to the three shortest (3- to 8-nt) products (Fig. 1, lanes 9–13). Thus, all potential sites were cleaved with high doses of the RNases, indicating full accessibility to the 3′ end of the RNA in both ECs. The action of the RNases was stopped with either phenol (lanes 2–7, 9–14) or urea/SDS (lane 15) (see Materials and Methods). Inactivation of the RNases with phenol, SDS, or other agents that denature proteins has been widely used in previous studies (9, 15, 25).
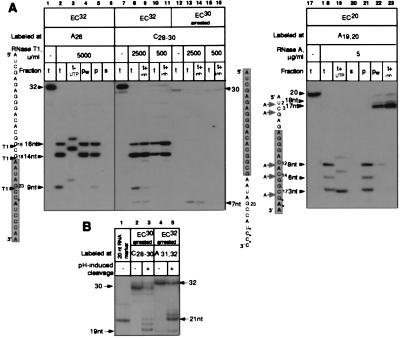
Using RNase as a tool for analyzing EC structure requires that all the cleavages occur in the RNA that forms a part of the intact complex. To ensure that the cleavage products did, in fact, originate from the intact ECs, we tested the integrity and catalytic activity of the digested complexes (Fig. 2A). To analyze the shortest 3′-proximal products of the cleavage, we selected high concentrations of the RNases: 5,000 units/ml RNase T1 and 5 μg/ml RNase A. All of the RNAs truncated by both nucleases, including the shortest 3- to 9-nt species, were quantitatively extended by the next nucleotide (UTP) (lanes 3 and 19). The elongation of the 3-nt RNA to the 4-nt product (EC20, lanes 18 and 19) was obscured by the abnormally low mobility of the GpApA trinucleotide on PAGE. Similar deviations in the migration of short nonphosphorylated oligonucleotides in denaturing PAGE were shown previously (26). The elongation of GpApA was confirmed by the digestion of the two gel-purified oligonucleotides with RNase T1 (data not shown).
Figure 2.
Analysis of the integrity and catalytic activity of ECs containing RNAs truncated with the RNases T1 and A. The origin of the cleavages introduced in the proximity of the RNA 3′ end. (A Left) Lanes 1–6, EC32 (lane 1) was treated with RNase T1 (lane 2, t, total). To test catalytic activity, the cleaved complex was incubated with UTP (lane 3, t+UTP). To test integrity, the cleaved complex was either washed (lane 4, pw, pellet washed) or divided into pellet and supernatant (lanes 5 and 6, p and s). Phenol was added to all the samples to inactivate the RNases. (A Center) Lanes 7–16, EC32 (lane 7) was treated with two doses of RNase T1. The cleavage was stopped either with phenol (lanes 8 and 10, t) or with 2′-GMP plus phenol (lanes 9 and 11, t+inh). Homogeneous arrested EC30 was obtained as described in Materials and Methods (lane 12) and treated with two doses of RNase T1. The cleavage was stopped either with phenol (lanes 13 and 15, t) or with 2′-GMP plus phenol (lanes 14 and 16, t+inh). (A Right) Lanes 17–23, EC20 (lane 17) was treated with RNase A and its catalytic activity and integrity were analyzed as described for A Left (lanes 18–22). Lane 23, the cleavage was stopped by Prime RNase Inhibitor plus phenol. The shaded rectangles represent the segments of the RNAs protected by RNAP in the intact ECs. (B) Homogeneous arrested EC30 and EC32 (lanes 2 and 4) were obtained as described in Materials and Methods and incubated at 37°C for 1 hr in TB adjusted to pH 9.0 by adding Tris base to stimulate the endonucleolytic activity of RNAP (27). Lane 1, 20-nt RNA used as a size marker.
An examination of the supernatant–pellet fractions obtained after a brief centrifugation of the digested ECs showed that all of the RNA products remained in the immobilized complex (compare lanes 2 and 5 and 18 and 21 in Fig. 2A). Furthermore, none of the products were detected in the supernatant fractions (lanes 6 and 20). At this point, all of the data were consistent with the view that the cleaved RNA was derived from intact ECs and was degraded to the minimal segments possible without apparent effects on the stability and activity of the complex. Unexpectedly, washing the truncated complexes with excess TB removed all RNAs shorter than 14 nt from the pellet (lanes 4 and 22). In addition, in EC20 the washing dramatically changed the pattern of the cleavage, causing the appearance of the longer 17-nt and 18-nt products (lanes 18 and 22).
Taken together, these results suggested that the RNases were not inactivated instantly after the addition of phenol and that the cleavage continued in the already denatured ECs. If the “postphenol” cuts generated the short 3- to 9-nt RNAs, this would explain the controversial properties of these products. The complete washing off of these fragments from the pellet could be attributed to the absence of the postphenol cleavage in the nuclease-free washed probe. The short products were not detected in the supernatant because no RNA was released from the EC at the moment the supernatant was collected. The apparent ability of the 3- to 9-nt products to be chased could be caused by postphenol degradation of the longer RNAs that were elongated before the phenol addition. To prevent the premature denaturation of ECs, we replaced phenol with nondenaturing inhibitors: 2′-GMP for RNase T1 and the Prime RNase Inhibitor for RNase A. As we expected, under the new conditions, all cleavages within 14 nt of the 3′-proximal RNA were eliminated in both complexes, matching all the effects of washing the samples with TB (Fig. 2A, compare lanes 8 and 9 and 18 and 23).
However, the 7-nt fragment generated by the RNase in the EC32 sample was not eliminated by 2′-GMP (lanes 8 and 9), which suggested that this cleavage occurred in the native complex. We observed the 7-nt RNA only in the EC32 sample that was labeled in the C28–C30 positions (compare lanes 2 and 8), when RNAP was stopped in position +30 during labeling (see Materials and Methods). Although most of the labeled RNA was extended to 32 nt after ATP addition, a small fraction of EC30 failed to resume elongation (lane 7). Because the RNA in this fraction did not dissociate from RNAP, this complex was considered to be arrested. We have shown previously that the loss of catalytic activity during the arrest is associated with the backward sliding of RNAP along the DNA and RNA, which makes the 3′ end of the transcript accessible to cleavage by the RNases (21). The 7-nt product could originate from the admixture of the arrested EC30. To prove that the arrested EC30 had retreated, we explored the ability of the RNAP active center to cleave the internal phosphodiester bonds in the RNA at pH 9.0 (27). At physiological conditions, this reaction is stimulated by the transcript cleavage factor GreB (22). We purified the inactive fraction of EC30 by prolonged incubation with a high concentration of four NTPs, which removed all elongation-competent complexes from the template (Fig. 2B, lane 2). The incubation of the arrested complex at pH 9.0 caused cleavage of the 30-nt RNA at 19 nt from the 3′ end, which positioned the active center in the arrested EC30 between A11 and C12 residues of the RNA (Fig. 2B, lane 3). As we expected, treatment of purified arrested EC30 with RNase T1 produced only the 7-nt fragment, whereas all cleavage sites at the 5′ end were protected as a result of the retreat (Fig. 2A, lanes 12–16). Similarly, the arrest of EC32 was responsible for the small 2′-GMP-insensitive fraction of the 9-nt product seen in lane 9 of Fig. 2A. In the purified arrested EC32 the pH-stimulated cleavage produced the 21-nt 3′ terminal RNA fragment, which confirmed the retreat of RNAP along the DNA and RNA (Fig. 2B, lanes 4 and 5).
Thus, in the active and intact EC20 and EC32, RNases A and T1 were unable to cleave the RNA closer than 14–17 nt from the 3′ end. Analogous experiments using the nondenaturing inhibitors were performed with other ECs (data not shown) and confirmed that all the cleavages detected within the 14-nt zone were introduced into the RNA that was released from RNAP. The previous accordant findings that the single-strand-specific RNases T1 and A were able to cut the nascent RNA 3–5 nt from the 3′ end provoked long-lasting debates about the length of the RNA–DNA hybrid in transcription (15). In these experiments, the protein-denaturing agents (urea and SDS) were employed to inactivate the RNases. Our present results argue that all these cleavages might also occur in the RNA dissociated from denatured RNAP rather than at the end of the RNA⋅DNA hybrid in the intact EC. In the denatured EC the transcription bubble must collapse and nontemplate DNA strand must force the RNA out of the hybrid. The relatively short RNA⋅DNA duplex is not strong enough to overcome the cooperative effect, which facilitates the two DNA strands’ rehybridization in the absence of RNAP. The denaturing agents themselves can also facilitate the RNA release from the duplex. We used two methods to avoid the artifactual postphenol cleavage of the released RNA: (i) applying low doses of the RNases and (ii) stopping the action of the high doses of RNases by using nondenaturing inhibitors or by washing the complexes.
Extrusion of the RNA from RNAP During RNA Chain Elongation.
To systematically investigate how the RNA translocates through RNAP in the course of transcription, we treated 15 ECs halted along the T7A1 transcription unit with high doses of RNases T1 and A. We used these high doses to digest secondary structures in the RNA; such structures might increase the apparent protection of the RNA in some complexes (28). The cleavage site closest to the 3′ end defined the upstream end of the protected zone. We washed the samples with TB to stop the action of the RNases. If the RNase was stopped by phenol, all potential sites in the RNA were cleaved. Washing prohibited the artifactual postphenol cleavage and removed all products derived from the RNA that was extruded from the enzyme during transcription arrest. The structural instability of some ECs (29) can produce additional problems. Washing allowed us to remove products originating from the spontaneously released RNA as well.
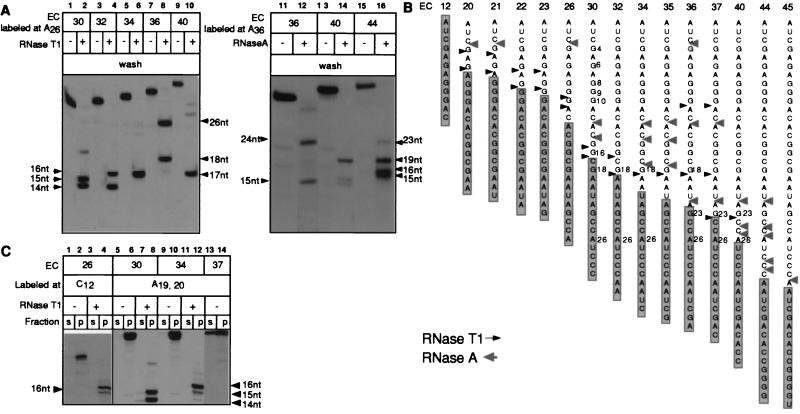
Fig. 3A Left shows five ECs halted 30–40 nt from the start point of transcription after treatment with 5,000 units/ml RNase T1 followed by washing with TB. The RNAs were labeled in the A26 position. The sequences of the RNAs are shown in Fig. 3B. Each of the original ECs contained 9 or 10 guanosines in their RNAs (marked G4–G35). In EC30, the RNA was cleaved efficiently at the G15 and G16 residues located 15 and 14 nt from the 3′ end (lane 2; cleavage at the G4–G10 sites was complete under these conditions). At the same time, the G18 site located at 12 nt was protected. The double products of the RNase cleavage reflect the difficulties for RNase to access the RNA at the very end of the protected zone. Advancing EC30 2 nt (to form EC32) translocated G18 to 14 nt from the 3′ end and made it accessible to RNase T1 (lane 4), whereas the cleavage at G15 was complete. This result showed that the extrusion of the RNA from RNAP occurred exactly 14 nt from the active center of the enzyme. Importantly, G23, located at 13 nt from the 3′ end, was totally protected in EC36 (lane 8), whereas its shift to 14 nt in EC37 (Fig. 3B) and further translocation to 17 nt in EC40 (Fig. 3A, lane 9) exposed G23 to the RNase. The presence of a 26-nt product in RNasetreated EC36 (lane 8) is explained by underdigestion of a very stable RNA hairpin that involves the G15 and G16 sites (29).
Figure 3.
Cleavage of the RNAs with RNases T1 and A in a number of consecutive ECs and their effect on EC stability. (A) The indicated ECs were digested with 5,000 units/ml of RNase T1 (Left) or with 5 μg/ml RNase A (Right), washed, and then combined with phenol. (B) Summary of RNase footprinting data obtained for the T7A1 transcription unit. Arrows show the cleavage sites introduced in the RNAs at high doses of RNases T1 and A, when the cleavage was stopped either by adding nondenaturing inhibitors or by washing off the RNases. The information shown is based on 3–5 separate experiments performed with RNAs labeled at various positions. The shaded rectangles represent the minimal 14-nt segment of the RNAs protected by RNAP in the intact ECs. (C) The indicated ECs, either intact or treated with RNase T1 and washed as described in A, were incubated in TB containing 300 mM KCl before separating the samples into supernatant and pellet (s and p).
The advancement of the 14- to 16-nt protected zone in the RNA accompanies elongation in other ECs as well. Lanes 11–16 of Fig. 3A illustrate this conclusion for EC36, EC40, and EC44 treated with RNase A. Fig. 3B summarizes all of the RNA cleavage data. DNA footprinting with exonuclease III has demonstrated that, in most of the complexes shown in Fig. 3B, RNAP translocated at 1 nt along the template as each new nucleotide was added to the RNA (30). The shaded box shows that the 14- to 16-nt protected zone advances in close synchrony with the growth of the RNA and with the translocation of the RNAP along the DNA. However, for some ECs, the translocation of the RNAP footprint on the DNA was shown to lag behind the growth of the RNA (30). This apparent delay in the enzyme’s translocation is explained by its back-and-forth oscillations along the nucleic acids at some positions in the template (23). In previous studies, protected regions of the RNA in oscillating ECs appeared slightly larger than those seen in normal complexes (23). In our experiments, the minimal 14- to 16-nt products were obtained even in the oscillating EC22, EC23, and EC26. The high doses of the RNases we applied were able to cleave the sites in the RNA behind the RNAP, which was covered temporarily by the oscillating enzyme.
Importantly, all the truncated products were retained in the pellet after washing with TB (Fig. 3 A and B) and were chased completely upon incubation with an appropriate NTP (data not shown). However, a decrease in the stability of the EC after RNA truncation might remain unnoticed at low ionic strength. Treatment with high salt has been widely used to evaluate the strength of the interactions holding the EC together (29, 31, 32). Therefore, we challenged the truncated complexes with high salt (300 mM KCl), which allowed us to discriminate between RNA that was tightly or loosely bound to RNAP. Under these conditions, the intrinsically unstable EC37 (29) partly released the RNA (Fig. 3C, lanes 13 and 14). At the same time, EC26, EC30, and EC34 did not dissociate, nor did their truncated versions (lanes 1–12). Thus, the RNA that was located more than 14–16 nt from the active center of RNAP was not required for the activity and stability of all the complexes tested. In the next section, we demonstrated that this remote part of the RNA dissociated after the cleavage.
Dissociation of the RNA Upstream from the Protected Zone.
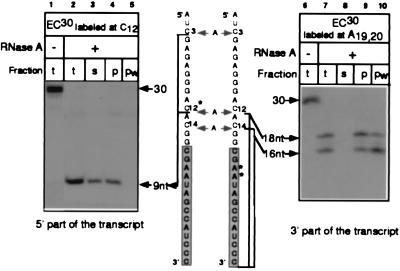
In our previous experiments (Figs. 1–3), labeling ECs near the growing ends of their RNAs did not allow elucidation of the fate of the 5′-proximal part of the RNA after the cleavage. Therefore, the RNA in EC30 labeled in C12 residue located upstream from the protected zone was treated with RNase A and the cleavage was stopped by the nondenaturing inhibitor (Fig. 4 Left). Cleavage at 18 and 27 nt from the 3′ end (after C3 and C12, respectively) generated the 9-nt labeled fragment (see the scheme in Fig. 4). This fragment was located next to the point at which the RNA was extruded from RNAP and contained most of the 5′-proximal part of the RNA. After cleavage, this fragment was harvested in the supernatant fraction (lanes 2–4) and could be completely washed off the complex (lane 5), indicating that it was not bound tightly to RNAP. A control experiment conducted with the same complex labeled within the protected zone demonstrated that the 3′-proximal part of the RNA was retained in RNAP under the same conditions and that the dissociation of its 5′ counterpart did not result from the disruption of the complex (lanes 6–10). Thus, the direct RNA dissociation assay strongly suggested that the RNA was freed from RNAP further upstream than 14–16 nt from the 3′ end.
Figure 4.
Analysis of the binding of RNAP to RNA upstream from the protected zone. The cleavage of EC30 with 5 μg/ml of RNase A was stopped with phenol in the presence of Prime RNase Inhibitor. Products of the cleavage (t) were either fractionated into supernatant and pellet (s and p) or washed with TB (pw) as described in Materials and Methods.
Isolation of Stable ECs Containing 8- to 10-nt RNA.
Elucidation of the minimal length of RNA required for EC stability is important in understanding the mechanism that triggers RNA release from the EC during transcription termination (2, 4, 13). Because the RNases failed to degrade the RNAs in the ECs to a length shorter than 14 nt, we further truncated these products from the 3′ end by using two known intrinsic ribonucleolytic activities of RNAP (see the scheme in Fig. 5). First, RNAP stimulated by the protein factor GreB hydrolyzes internal phosphodiester bonds in the nascent RNA (22). Second, in the presence of PPi, the RNA can be shortened progressively by a reversal of the NTP-addition reaction (33). The treatment of RNase-truncated EC26, EC30, and EC34 with GreB or PPi produced the 8- to 10-nt RNAs (Fig. 5). These RNAs did not dissociate from RNAP in high salt and could be elongated with the appropriate NTPs. Using PPi, we ruled out the possibility that binding of GreB to RNAP might stabilize the complexes. Thus, the 8- to 10-nt region of the transcript was sufficient to stably anchor of the RNA to RNAP in these ECs.
Figure 5.
Shortening of RNase-cleaved transcripts with GreB or PPi and analysis of the stability and catalytic activity of the resulting ECs. The scheme on the left illustrates the format of the experiment. ECs treated with RNase T1 were incubated with GreB or PPi and tested either for their stability in TB containing 300 mM KCl followed by separation into supernatant and pellet fractions (s and p) or for their catalytic activity (t+NTP). The sequence at the bottom shows the truncated products that were obtained.
The retreat of RNAP caused by factor-induced cleavage of the RNA, pyrophosphorolysis, or transcription arrest is associated with a shift of the transcription bubble and with reverse-threading of the RNA through the enzyme (21, 26). In terms of nucleic acids, this retreat can be visualized as a backward translocation of the RNA⋅DNA hybrid (see the scheme in Fig. 5). For this process to be energetically justified, the disruption of one RNA–DNA pair at the leading edge of the transcription bubble must be compensated by the formation of one RNA⋅DNA base pair at the rear end of the hybrid. We proposed earlier that the 5′ end of the RNA blocks reverse translocation of RNAP because RNA⋅DNA rehybridization at the upstream RNA–DNA junction no longer persists (21). These considerations suggest that in the experiment described above, the RNAs were reduced by GreB or PPi to their part involved in the hybrid with the DNA. Also, recent results obtained by RNA–DNA crosslinking and by assembly of functional ECs from synthetic nucleic acid components have shown that the 8- to 9-nt region of the 3′-terminal RNA forms a hybrid with the template (14, 32). Thus, the data in Fig. 5 indicate that the interactions between RNAP and its transcript that are crucial for the EC stability are restricted to the part of the RNA that is hybridized to the DNA.
In this work, the effect of truncation of the RNA to 8 nt was analyzed in a single template position. Our assembly experiments showed that the overall strength of the RNA⋅DNA hybrid also contributes to the stability of EC (32). Therefore, we expect that the stability of ECs containing 8-nt RNA may vary from one template position to another, depending on the strength of the local RNA⋅DNA hybrid.
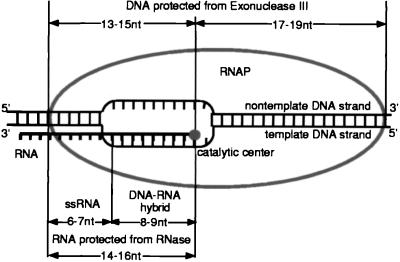
Model for Functional Topography of Nascent RNA in the EC.
The results of our studies are summarized as follows. (i) The minimal length of the RNA produced by treatment of 15 intact consecutive ECs with the highest doses of RNases A and T1 was consistently 14–16 nt. (ii) All of the cuts within the 14- to 16-nt region occurred either in the complexes in which the 3′-proximal RNA was extruded from the protein as a result of transcription arrest or in the free RNA released from the denatured EC during RNase inactivation. (iii) The 14- to 16-nt 3′-proximal fragment was sufficient for the stability and activity of all RNase-treated complexes regardless of the initial length of the RNA. The remaining transcript rapidly dissociated from RNAP. (iv) GreB- or PPi-induced shortening of the 14- to 16-nt RNAs to 8–10 nt did not affect the stability and catalytic activity of the four ECs that we tested.
Fig. 6 represents the RNA organization in the EC based on our current results and on the data obtained previously (11, 14, 21, 23, 25). The 8–9 nt of the 3′-proximal RNA form a hybrid with the template as follows from the RNA–DNA crosslinking experiments in which the RNA was crosslinked to the template DNA up to 8–9 nt from the 3′ end (14). The use of a novel reconstitution technique for the assembly of active EC from the short synthetic RNA and DNA oligonucleotides led us to the same conclusion, because a 9-bp RNA⋅DNA hybrid was required for the formation of stable EC (32). Our current finding that proper inactivation of RNases is required to avoid cleavage of the RNA in denatured ECs contradicts the theory of a short RNA⋅DNA hybrid in the EC based on the apparent sensitivity of the nascent RNA to the single-strand-specific RNases 3 nt from the 3′ end (15, 16).
Figure 6.
The RNA organization in the active EC. See the text for details.
The 14–16 nt in the RNA are protected from the RNases in the EC. We believe that this protection is caused by the passive block imposed by RNAP. Protection by a 14- to 16-nt RNA⋅DNA hybrid is not supported by the bulk of the experimental data cited above. Our results argue that the rest of the RNA upstream from the protected zone is dispensable and is extruded from the enzyme. On the other hand, the crosslinking of the RNA to RNAP at distances greater than 14 nt from the 3′ end favors the theory of an extended RNA-binding site in RNAP (11). However, these data can be explained either by the nonspecific interactions of the RNA with charged patches on RNAP or by the transcript’s ability to form a secondary structure that could bring the remote, crosslinkable group close to the surface of the protein.
We assume that the 8- to 10-nt region that forms a hybrid with the DNA determines the overall stability of the EC (this work and ref. 32). The next 6–7 nt of the RNA, although covered by RNAP, are not required for the stability and activity of the complex. In vitro studies of transcription initiation at rrnB P1 and λPL promoters and studies of ECs obtained on short synthetic DNA oligonucleotides using template switching revealed extremely stable complexes containing only 4–6 nt in the RNA hybrid, demonstrating that the short hybrid may be enough to stabilize the EC (18, 19, 31). However, the promoter complexes resulted from the unusual reaction of RNA slippage, whereas the ECs obtained by the template switching contained no DNA upstream from the RNA⋅DNA hybrid and had no noncoding DNA strand in the region of the transcription bubble. Moreover, the complex derived from rrnB P1 contained the σ subunit. Thus, the stabilization mechanism of these complexes may be different from that used in normal elongation.
Nevertheless, in template positions in which the 3′-proximal RNA⋅DNA pairing is unstable, ECs containing RNA longer than 9 nt may escape dissociation by the temporary retreat of RNAP to a more stable hybrid. Because of the multiple hybridization choices available, the average stability of these ECs may be higher than that of the complexes with the 9-nt RNAs. This search may be restricted by the secondary structures in the RNA that stop RNAP retreat, which may explain the destabilization role of the RNA hairpins in transcription (29).
Acknowledgments
This work was initiated while N.K. and M.K. were at Dr. A. Goldfarb’s laboratory in the Public Health Research Institute (New York). We thank Dr. Goldfarb for generous support and advice. We are grateful to Dr. M. Kireeva and I. Sidorenkov for helpful discussion and to I. Sidorenkov for purification of RNAP and GreB. This work was sponsored by the National Cancer Institute, Department of Health and Human Services, under contract with Advanced BioScience Laboratories and by National Institutes of Health Grant GM 49242 (to Dr. Goldfarb).
ABBREVIATIONS
- RNAP
RNA polymerase
- EC
elongation complex
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Uptain S M, Kane C M, Chamberlin M J. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 2.Yager T D, von Hippel P. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1987. pp. 1241–1275. [Google Scholar]
- 3.Richardson J P, Greenblatt J. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd Ed. Neidhardt F C, Curtiss R, Ingraham J L, Lin E C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 822–848. [Google Scholar]
- 4.Landick R. Cell. 1997;88:741–744. doi: 10.1016/s0092-8674(00)81919-4. [DOI] [PubMed] [Google Scholar]
- 5.Das A. Annu Rev Biochem. 1993;62:893–930. doi: 10.1146/annurev.bi.62.070193.004333. [DOI] [PubMed] [Google Scholar]
- 6.Chan C L, Landick R. In: Transcription: Mechanisms and Regulation. Conaway R C, Conaway J W, editors. New York: Raven; 1994. pp. 297–321. [Google Scholar]
- 7.Kumar S A, Krakow J S. J Biol Chem. 1975;250:2878–2884. [PubMed] [Google Scholar]
- 8.Lee D N, Landick R. J Mol Biol. 1992;228:759–777. doi: 10.1016/0022-2836(92)90862-e. [DOI] [PubMed] [Google Scholar]
- 9.Gu W, Wind M, Reines D. Proc Natl Acad Sci USA. 1996;93:6935–6940. doi: 10.1073/pnas.93.14.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monforte J A, Kahn J D, Hearst J E. Biochemistry. 1990;29:7882–7890. doi: 10.1021/bi00486a015. [DOI] [PubMed] [Google Scholar]
- 11.Hanna M M, Meares C F. Proc Natl Acad Sci USA. 1983;80:4238–4242. doi: 10.1073/pnas.80.14.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gamper H B, Hearst J E. Cell. 1982;29:81–90. doi: 10.1016/0092-8674(82)90092-7. [DOI] [PubMed] [Google Scholar]
- 13.Yager T D, von Hippel P. Biochemistry. 1991;30:1097–1118. doi: 10.1021/bi00218a032. [DOI] [PubMed] [Google Scholar]
- 14.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 15.Rice G A, Kane C M, Chamberlin M J. Proc Natl Acad Sci USA. 1991;88:4245–4249. doi: 10.1073/pnas.88.10.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chamberlin M J. Harvey Lect. 1994;88:1–21. [PubMed] [Google Scholar]
- 17.Altmann C R, Solow-Cordero D E, Chamberlin M J. Proc Natl Acad Sci USA. 1994;91:3784–3788. doi: 10.1073/pnas.91.9.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Severinov K, Goldfarb A. J Biol Chem. 1994;269:31701–31705. [PubMed] [Google Scholar]
- 19.Borukhov S, Sagitov V, Josaitis C A, Gourse R L, Goldfarb A. J Biol Chem. 1993;268:23477–23482. [PubMed] [Google Scholar]
- 20.Kashlev M, Nudler E, Severinov K, Borukhov S, Komissarova N, Goldfarb A. Methods Enzymol. 1996;274:326–333. doi: 10.1016/s0076-6879(96)74028-4. [DOI] [PubMed] [Google Scholar]
- 21.Komissarova N, Kashlev M. Proc Natl Acad Sci USA. 1997;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borukhov S, Sagitov V, Goldfarb A. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- 23.Komissarova N, Kashlev M. J Biol Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 24.Kashlev M, Martin E, Polyakov A, Severinov K, Nikiforov V, Goldfarb A. Gene. 1993;130:9–14. doi: 10.1016/0378-1119(93)90340-9. [DOI] [PubMed] [Google Scholar]
- 25.Wang D, Meier T I, Chan C L, Feng G, Lee D N, Landick R. Cell. 1995;81:341–350. doi: 10.1016/0092-8674(95)90387-9. [DOI] [PubMed] [Google Scholar]
- 26.Feng G H, Lee D N, Wang D, Chan C L, Landick R. J Biol Chem. 1994;269:22282–22294. [PubMed] [Google Scholar]
- 27.Orlova M, Newlands J, Das A, Goldfarb A, Borukhov S. Proc Natl Acad Sci USA. 1995;92:4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yakovlev G, Moiseyev G P, Sorrentino S, De Prisco R, Libonati M. J Biomol Struct Dyn. 1997;15:243–250. doi: 10.1080/07391102.1997.10508189. [DOI] [PubMed] [Google Scholar]
- 29.Arndt K M, Chamberlin M J. J Mol Biol. 1990;213:79–108. doi: 10.1016/S0022-2836(05)80123-8. [DOI] [PubMed] [Google Scholar]
- 30.Nudler E, Goldfarb A, Kashlev M. Science. 1994;265:793–796. doi: 10.1126/science.8047884. [DOI] [PubMed] [Google Scholar]
- 31.Nudler E, Avetissova E, Markovtsov V, Goldfarb A. Science. 1996;273:211–217. doi: 10.1126/science.273.5272.211. [DOI] [PubMed] [Google Scholar]
- 32.Sidorenkov I, Komissarova N, Kashlev M. Mol Cell. 1998;2:55–64. doi: 10.1016/s1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- 33.Rozovskaya T A, Chenchik A A, Beabealashvilli R S. FEBS Lett. 1982;137:100–104. doi: 10.1016/0014-5793(82)80323-2. [DOI] [PubMed] [Google Scholar]